

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
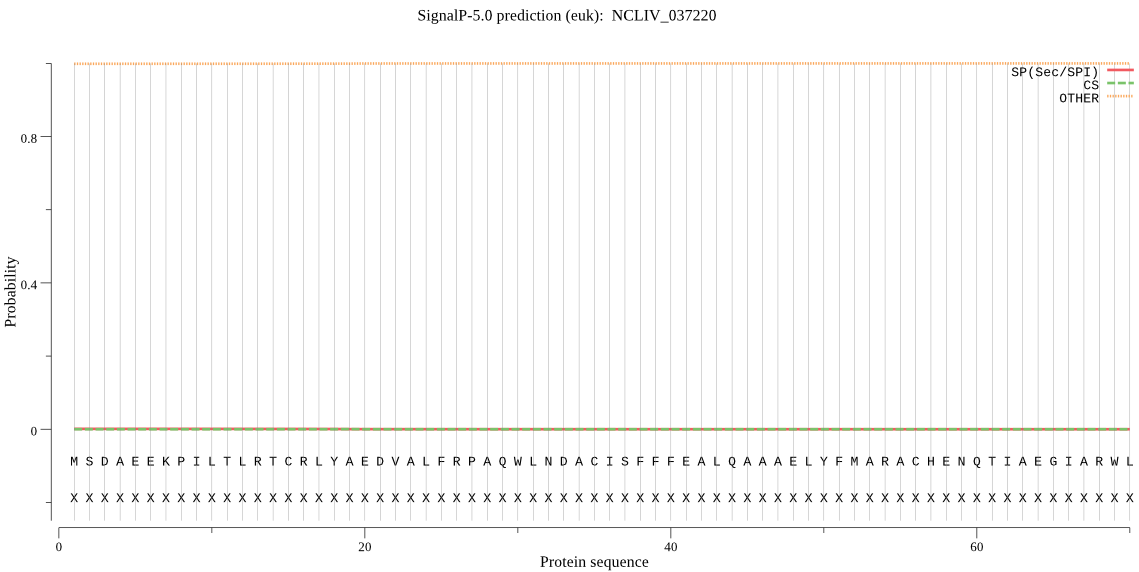
| NCLIV_037220 | OTHER | 0.999838 | 0.000150 | 0.000012 |
MSDAEEKPILTLRTCRLYAEDVALFRPAQWLNDACISFFFEALQAAAELYFMARACHENQ TIAEGIARWLAEGIEPLSEKNEELLRCKLGLSSPACQPSAGEPASCNVTSRQEEHSEALL SLDLTEQDEAQVCSPILFVDPCVAFWVSMCTDESEVQDTLENLHIPRRSVVVWPITDHQD PSTVGGTHWTLLIQIVCNRAVNRNATAQNDYNAHDPVQLLSGYQYFHFDSIGHNCQQSRG NLAAARRLQALLGRFVKISTSTDSPPAGDHDRSDRTPIPLRCTPPQSNASDCGVHCLLFA ERLLQLVVKKDRDFFKGKDNSGDRRSKQTTPSVDVETGPLYPELPFDVSWFSLPDLQRFE PSVLAARRRGAILLVNKLTQRGGGC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_037220 | 78 S | IEPLSEKNE | 0.996 | unsp | NCLIV_037220 | 264 S | TSTDSPPAG | 0.993 | unsp |