

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
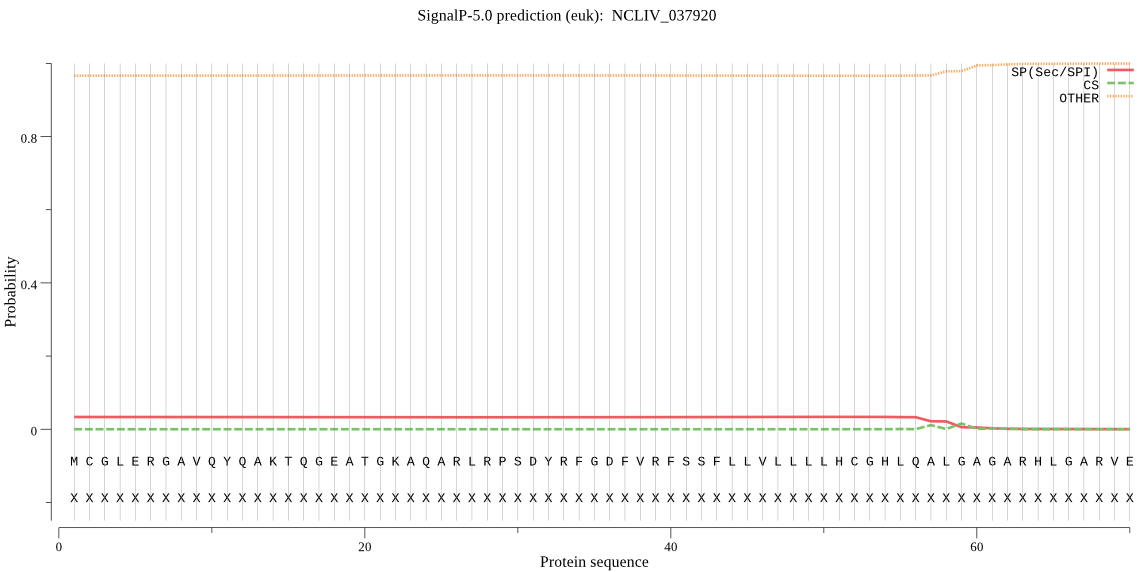
| NCLIV_037920 | OTHER | 0.996008 | 0.001998 | 0.001994 |
MCGLERGAVQYQAKTQGEATGKAQARLRPSDYRFGDFVRFSSFLLVLLLLHCGHLQALGA GARHLGARVEAARSFWRGVDAARKRSRIRTRDSSKSPPSFLTLSPFPSKPASRLPGVHSD GHPLSFLQPPADVPRPFSGLPRAEERRKGTRPSLKTDVLATQRRLVSRLPPRHSVRRSHL LAARSVSDAYLTISPRASPHPNLSLKFTRDPTDRMPFRSLGAAQRPSAGGLELAVTDGEK ERDGGAHASSSRSDKQDVPTTTAVPFFAFGESEPATFLADERETRAPEASPEPQRRVFPD LPTADIRSAMAHSLPQNENAPPRETLLSPRSRQSASATASSLPFAPSEERHPRATLLAYV TGFGPFGSVRKNPTACLVSHMERALLRAQDGEAGFEEQRMPSPYSSSEGSMRSGQPANAA EFGFFVQSGVSFEDLATSCAADDGAASSPKSGKQEGSIVGGSTRRNVADINSDGTGSPEA LVAEISRSAKDGVFAADKARSQSGEETVITRAFTNPPPVHFISSGVRLCGAEILEAAAAA AKEAAPRIGAALRLNSSWTERQTRLRSLDVRDSARLQAFGAAQESGVKTPTVSPSAVAKN GIRDRLGKTLAEVESSETQRLGDTDSPVRMRGLPEAASPVQADTPMVEAAASEKNGSQGK PGDEHETEADEKRVVKKLAFHLGLNQAATAFELEKVAVNEADFCIPDQRGFRPEKARISD TGPDRLVTRLPLEEICTALQERGFPCKTSTYAGRFVCNYLYYQSLLENLGSDTEVLFVHV PPFSTIPYSWQVSFLLQLLDVIRRLPEAPLTTSADAARNKKQHARRTSKASGSSGGNPKV EKEPEYEVEDVVAYTEDRRGVARWKVRWKGYDSDEDTWETRANLRGSPAFWKQMDRLQRE WRMAHKNDESSSAFEDEDEDDGRSEDDVADPTRPGKGQSKARRGAGRPPAGKRRRTHSSS KRGKASSEDENEEEHEGSEEAPGVEDADREQDSSGEDDELLPYPIIYGTDTEPPPHHLPD ETVLTDGRLSVFRFKNVPLKPKSDAHLGDTPACEGLSPGRLPRTEREDGSGLPAKRERRS AETENVRLSGRPGGDAAGGEDSKTAPEAQPASAHGDAPSKGGNCVYVQYLVDGKYVYSLP FDRARLYCPQLLLTYLMARTTFKTSAAAEGRPSRGKTRQAYESCFVQENQENLAREANCL PSPERGSVTPASVEEVEVDNAVAEEKLYSDAAVLSRETTSSTGRKRDGGHVDEESERTDS GDESESHAEKAHTRQKAGNRFPEEATRRDSSETDGSEFASDGSDSGGSKSGQAKKDDAGG AVEYEYGALKRSPIFSCVAG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_037920 | 94 S | TRDSSKSPP | 0.996 | unsp | NCLIV_037920 | 94 S | TRDSSKSPP | 0.996 | unsp | NCLIV_037920 | 94 S | TRDSSKSPP | 0.996 | unsp | NCLIV_037920 | 96 S | DSSKSPPSF | 0.991 | unsp | NCLIV_037920 | 153 S | GTRPSLKTD | 0.992 | unsp | NCLIV_037920 | 174 S | PPRHSVRRS | 0.996 | unsp | NCLIV_037920 | 198 S | SPRASPHPN | 0.996 | unsp | NCLIV_037920 | 253 S | SSSRSDKQD | 0.996 | unsp | NCLIV_037920 | 290 S | APEASPEPQ | 0.996 | unsp | NCLIV_037920 | 334 S | RSRQSASAT | 0.993 | unsp | NCLIV_037920 | 402 S | QRMPSPYSS | 0.996 | unsp | NCLIV_037920 | 405 S | PSPYSSSEG | 0.992 | unsp | NCLIV_037920 | 431 S | QSGVSFEDL | 0.996 | unsp | NCLIV_037920 | 448 S | GAASSPKSG | 0.997 | unsp | NCLIV_037920 | 451 S | SSPKSGKQE | 0.996 | unsp | NCLIV_037920 | 477 S | DGTGSPEAL | 0.991 | unsp | NCLIV_037920 | 488 S | EISRSAKDG | 0.997 | unsp | NCLIV_037920 | 501 S | DKARSQSGE | 0.992 | unsp | NCLIV_037920 | 503 S | ARSQSGEET | 0.998 | unsp | NCLIV_037920 | 719 S | KARISDTGP | 0.994 | unsp | NCLIV_037920 | 873 S | KGYDSDEDT | 0.995 | unsp | NCLIV_037920 | 959 S | RTHSSSKRG | 0.996 | unsp | NCLIV_037920 | 966 S | RGKASSEDE | 0.996 | unsp | NCLIV_037920 | 967 S | GKASSEDEN | 0.997 | unsp | NCLIV_037920 | 993 S | REQDSSGED | 0.996 | unsp | NCLIV_037920 | 994 S | EQDSSGEDD | 0.998 | unsp | NCLIV_037920 | 1030 S | DGRLSVFRF | 0.996 | unsp | NCLIV_037920 | 1080 S | RERRSAETE | 0.993 | unsp | NCLIV_037920 | 1089 S | NVRLSGRPG | 0.996 | unsp | NCLIV_037920 | 1173 S | EGRPSRGKT | 0.996 | unsp | NCLIV_037920 | 1202 S | NCLPSPERG | 0.994 | unsp | NCLIV_037920 | 1212 S | VTPASVEEV | 0.996 | unsp | NCLIV_037920 | 86 S | ARKRSRIRT | 0.996 | unsp | NCLIV_037920 | 1260 S | ERTDSGDES | 0.998 | unsp | NCLIV_037920 | 1290 S | TRRDSSETD | 0.998 | unsp | NCLIV_037920 | 1291 S | RRDSSETDG | 0.998 | unsp | NCLIV_037920 | 93 S | RTRDSSKSP | 0.998 | unsp |