

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
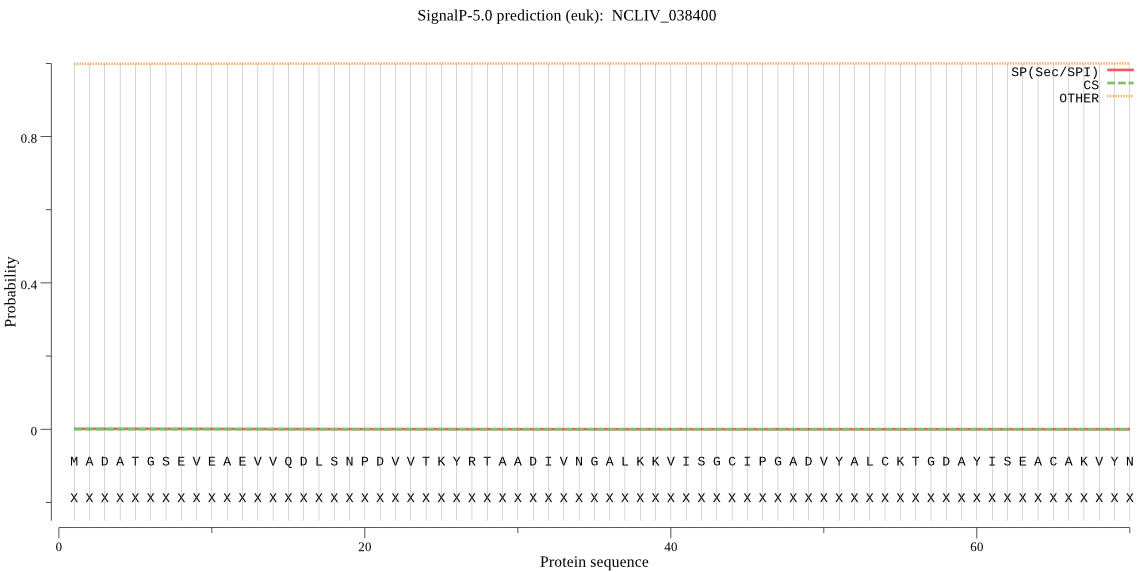
| NCLIV_038400 | OTHER | 0.999835 | 0.000161 | 0.000004 |
MADATGSEVEAEVVQDLSNPDVVTKYRTAADIVNGALKKVISGCIPGADVYALCKTGDAY ISEACAKVYNKKENGKKMEKGIAFPTCISINEICGHFSPVEENAETDRVLTEGDVVKIDM GCHIDGYISVVAYTVVCDAALPGIFEGTPGAQQGRISGRKADVIKACWTAAEACMRLVKA GHKSTDLTKTIELAAKQYGCTPLQGVLSHQLKRYVIEGSKCFAGATPGPGEDEPEEFEFE PNEVYGVDIVISSGEGKARDAAIKPTVYKRAVDRTYILKSQLGRHFMSEVQNKYPTLPFS LRGFSDDRACKVGVAEAMRHELLHPYPVMTEKPGEFVAQFKFTLLLLPTGTKKVTGLPLL YEKELDSSHTVEDESLKALLAVSVNPKKLKKKAQVEKKEDGGNA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_038400 | 157 S | QGRISGRKA | 0.996 | unsp | NCLIV_038400 | 375 S | VEDESLKAL | 0.991 | unsp |