

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
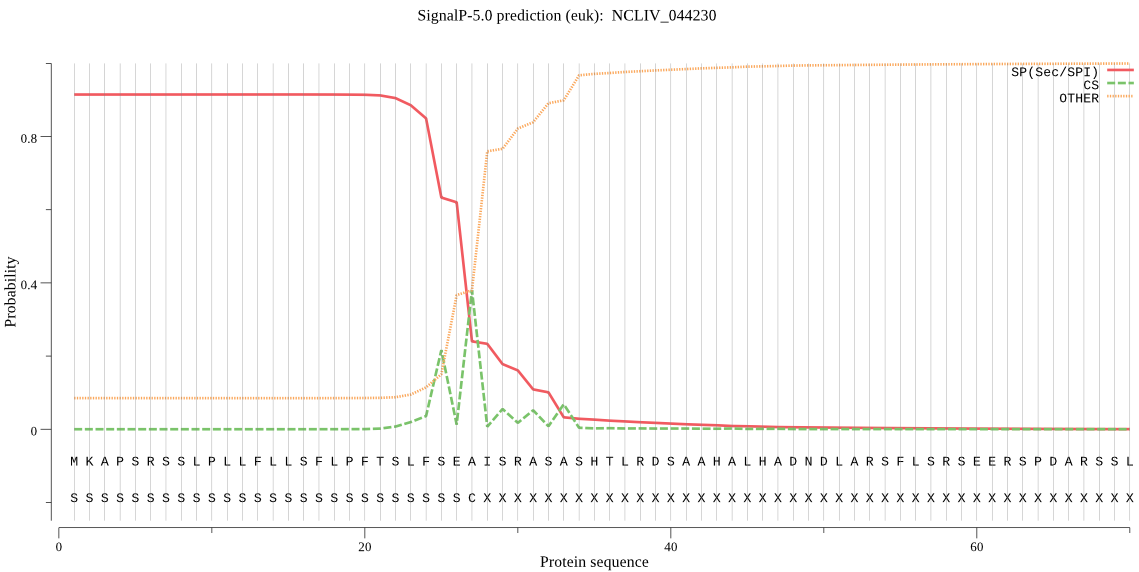
| NCLIV_044230 | SP | 0.156806 | 0.843085 | 0.000109 | CS pos: 27-28. SEA-IS. Pr: 0.3230 |
MKAPSRSSLPLLFLLSFLPFTSLFSEAISRASASHTLRDSAAHALHADNDLARSFLSRSE ERSPDARSSLEEEADAVENASSHLPDLEDALQQELESASSSPETVPLSSSEGASDPSPSS FVSSSPTSRASAASASSREAPDSLPTLAEVHASLASRIAQERSSRRLSDAEHGEDANDEN GDRKHVRKPPRDKSAYSVFSVPSLRLEGVAIADKEEATASFAISVGCGFFHDPPAIPGIA HQLEHLIFLGAEGETNATSWDEFVSARGGTHNAHTTAELTTFFVAAPTDTLPELLDRFLQ HLFRPLLAADQFASEVMAVQFEHEKNQPDIARVLLELAMAATPAVGSPTSTSREDLPPSF YRPEVAQKFGTGDYETLCQAPIEQGLDVLKALRAFHRKCYRPENMTIAVRMGRRSVPVVI EGSFDSAGRPRPIPAEVQAEAERQAEKYSVYTPKEIGEVVARILTKYFPPTEEAESDAHR ADNEDAGASPSDPPSASLTPRPHKASSSASSASRSGAKAKASTTADAKRRKAVLKTVRLH AEEKEKTGAQDAASFLEVTPEEHASTLEDDTELAEEDSIQADATSSTSTGKVYRVFRQHG WGKRLLLVWERRTSWAVRGFDEEFQPTALLEYLLEYPGETALLNRLKVQGLIADGEYVDY TTSQKAFIGLLFELTDEGEKNHEQVIAATLAYADTLRTSVDDAYILDFFEEFAGVANRSW TYKDPEDAVSAVIGAAEKLAVFPQRPDLVVAGGEYVALPEDRSVLVRVLREELQSFDSSR ASAFVILPEGRARDAGQDVHVFAPYAVQYTVTELPAPEVARQVGAKIGRSAAEKLEAVRE AAESVQLRTQSLVSVQETVFSLPPVLSCKLAAEVKLQRRPVPDVCQSLSPIQFPIQAERN EGEGAAKRAGFSTGARAGVRDADAEDDDNEQKKLEASGPPCVLVTEESFSVFWKNAEPFN KPVVRAYFKLRISAEDATGKNTLYGKIFTTIAGERARTALATFEGCGVDLLMSFTNGALV LEIQARKETFRAFSELFASVLQQLIAVLKETRESVKEADFTKVFNTLELQLSDFSAVTPF EMAMDVALSVIRRNRFSQLDLRRAATESSSASQFEEFQNFVRNALTRNALDVFIMGDIDY DAARELAEEFRAALSEQPLPFAQSAGNEILNVAEDIEIRFPNPIPEDATNAYVSLYVTHP PPDMMEMVIYSLIGEVISSPFFDTIRTHWMDGYVAAAAVREVPPAMTLATIVQGSKRKPD ELERHVCAFLAEMQDNIGTSMTTETFLERLRWLSSSKFHRSATSFSDYFGEVTSQIASRN FCFIREQLARLATEQFLNCPAILKHYMNALVDRANRKRIAVKIEGNTPSAEAASGSTRAT ENQPLSSAALPSRCSLPSLSSSASASAEVRQSPSLSTLTPAVVPRAVAKHDGRSQRETET SESSSSSANVKGQAKTGDQTRTSSASSLGSSHKDGAEAEHLAGRVVSLLGVGEESRGAKA PVNERRSYGSLLGSHNVFKSNRIKTDFASPIFTLGDDEEALSLIQVDARISTQSEHAKPV LGSRKAEGHSQAPSAAVAEPSASKNRKGRSQEPTETPGLEITEIVSDVEGQAEEGDVPIE FLPLAKQIMPHILERCEEDQKAAADGKRIVVDNFFTDPDEARRVILEKLGAKLNSSGSAP PTAAAASPLWLSYTRAESCSIRTSTIAKAEEVCGSIREAEMDAVPLVSPEQPSVGQPSIP PTLAAVPPTSEEAPPSAAAAGAAGAAQEPDQAARAADLEDASAAETEELRLRQSAKQRRL EQEKSLLEARLTRLELQTIAQRAQLLKQRLDAMKQYGTRPTTIFNTWLAPQRRGIRDVGG GKGASSGAFPVAAVSALGATTHGLPSPQGVQGRLRSPSSLHHAADPLAFKRTLDTLRWRE QALLQRQAELRNQLRAEPPSNASTPASTVPSWGAAPGQAIRPVEYVAAPAVASVPSSPYL GSSSRTIAANQAISVPATTAPNAPPAVAASPVAASPAVAASPVAASPVAVAAVPQIPEQV PGLGVGVAAPGLSSVSGVSAAPSPALAAPVSAAAPVPAAAPVPAAAPVSAAAPVPAAAPV SAAAPVSAAAPVSAAAPVSAAAPVSAPSPALAAPVSAAAPLALSPAFQAGLAATAPAGGE LSESPLVQATVSGVPRVVAAAANFQAPAAQPAVASQPPSPQAVSPGAPAPVAAGASATPA VIQPTAPVAAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_044230 | 100 S | ESASSSPET | 0.99 | unsp | NCLIV_044230 | 100 S | ESASSSPET | 0.99 | unsp | NCLIV_044230 | 100 S | ESASSSPET | 0.99 | unsp | NCLIV_044230 | 108 S | TVPLSSSEG | 0.993 | unsp | NCLIV_044230 | 136 S | AASASSREA | 0.994 | unsp | NCLIV_044230 | 137 S | ASASSREAP | 0.994 | unsp | NCLIV_044230 | 164 S | QERSSRRLS | 0.997 | unsp | NCLIV_044230 | 168 S | SRRLSDAEH | 0.998 | unsp | NCLIV_044230 | 351 T | SPTSTSRED | 0.992 | unsp | NCLIV_044230 | 352 S | PTSTSREDL | 0.997 | unsp | NCLIV_044230 | 489 S | DAGASPSDP | 0.997 | unsp | NCLIV_044230 | 511 S | SSASSASRS | 0.993 | unsp | NCLIV_044230 | 699 S | TLRTSVDDA | 0.998 | unsp | NCLIV_044230 | 854 S | QSLVSVQET | 0.996 | unsp | NCLIV_044230 | 973 S | KLRISAEDA | 0.998 | unsp | NCLIV_044230 | 1054 S | ETRESVKEA | 0.998 | unsp | NCLIV_044230 | 1097 S | RNRFSQLDL | 0.997 | unsp | NCLIV_044230 | 1112 S | SSSASQFEE | 0.992 | unsp | NCLIV_044230 | 1304 S | RSATSFSDY | 0.99 | unsp | NCLIV_044230 | 1412 S | EVRQSPSLS | 0.992 | unsp | NCLIV_044230 | 1434 S | HDGRSQRET | 0.998 | unsp | NCLIV_044230 | 1445 S | SESSSSSAN | 0.992 | unsp | NCLIV_044230 | 1467 S | SSASSLGSS | 0.991 | unsp | NCLIV_044230 | 1471 S | SLGSSHKDG | 0.998 | unsp | NCLIV_044230 | 1590 S | RKGRSQEPT | 0.994 | unsp | NCLIV_044230 | 1750 S | VPPTSEEAP | 0.993 | unsp | NCLIV_044230 | 59 S | FLSRSEERS | 0.994 | unsp | NCLIV_044230 | 69 S | DARSSLEEE | 0.998 | unsp |