

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
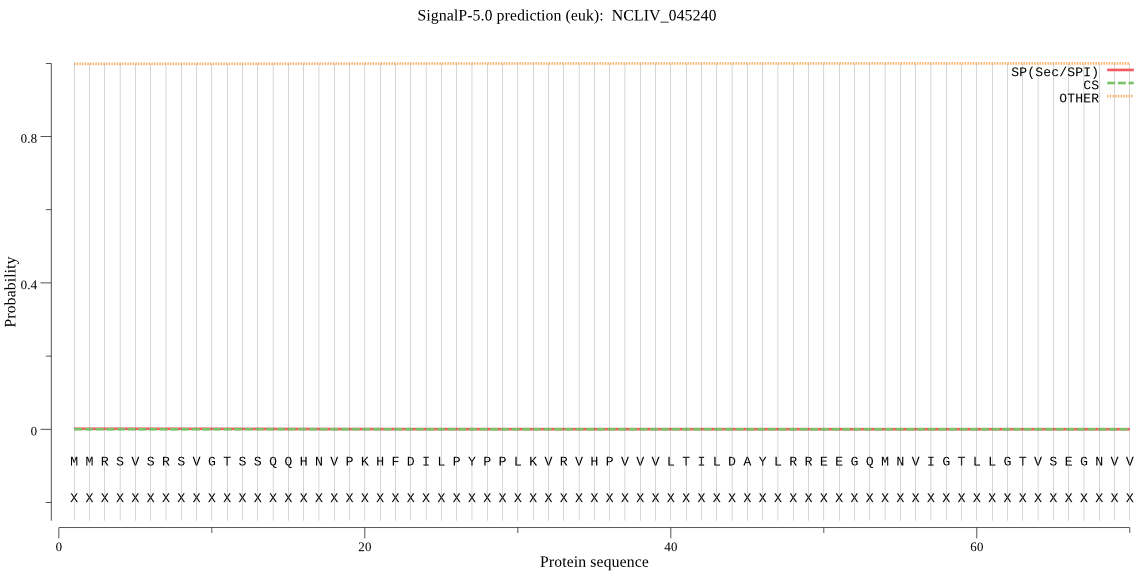
| NCLIV_045240 | OTHER | 0.889000 | 0.001463 | 0.109538 |
MMRSVSRSVGTSSQQHNVPKHFDILPYPPLKVRVHPVVVLTILDAYLRREEGQMNVIGTL LGTVSEGNVVDISDCFVDRHSLTDEGLLQIIKDHHETMYELKQQVSGSGAKDIVVGWFCT GSEMTELTCAVHGWFKQFNTVSKFHPQPPLTEPIHLMVNTTMDRDNLSIKAYMQVPMNMA KDACFQFQELPLELFASSSDRAGLSLLLKVREANRRHRQQHEGNRASALSSSAPSSAAAV AAISDTPGVAGVPVIKQGLGAALEKLSDQLNKCGAYVRSVLDGSEKADPEIGRFLSKALC VEAVQDLEVFEQMCQNALQDNLMVVHLTSLARLQFAVAEKLNTSFF
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_045240 | 81 S | VDRHSLTDE | 0.998 | unsp |