

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
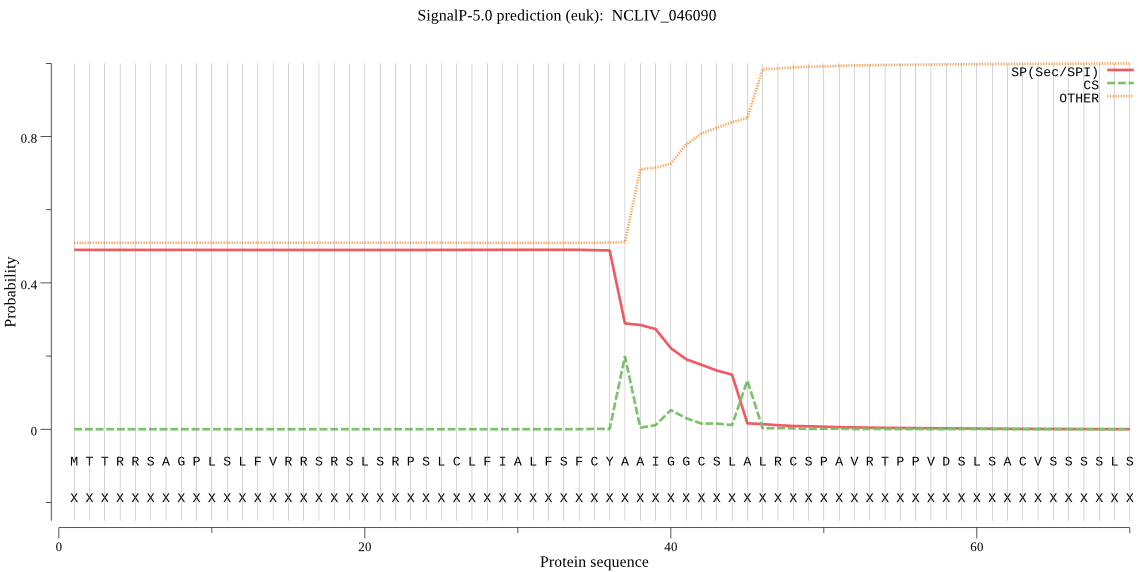
| NCLIV_046090 | OTHER | 0.773225 | 0.212487 | 0.014288 |
MTTRRSAGPLSLFVRRSRSLSRPSLCLFIALFSFCYAAIGGCSLALRCSPAVRTPPVDSL SACVSSSSLSPSAAPGGLSSPVAVRCSASSRALCSASAPRRSSALCCSHPHDGGTENAKE PRTRDLHALLLQAAAVEDSKVLLLLESAARLYANPESSRRLRERNRGRRRESRIKQGRTG RPDGNERRRAEQSDRALRARDTHFEDAFHPENAVLGLSPGDGHEVERPRVENETRTGGKG NATSTDSAPELPKGFLPLLFSRNARFANTGVPTVFRPLPPFLSLRRDRGRAGREDTEGKR DDAREDDCEGETEGAGDEDVPPFLHRLPPDALQKQLRERRKWKPPSAVRALLENPRVDQT GNEPNASFSVHSSSGSSPSSFASSCPSHAFSPACDASAVVASSVSSSSPELPGAASSLAS AASEDRPSGRIRGRRGVGPRVIEATAILARVVKCESCSRGDSSQERWHAFPASGVFIHPS GILVTNFHVLDSSQDRADADGEQEGTAEPEGEREREEREEEEQEEEARGGKESGAQVVEK SQDTSGKMDQRRGGGETERWGGAFDKARGGADRRGTARQKSRHRKRSFTQREEMFIVMDR RGRCWPVQEVLAADEETDLALLKVDWDGLVDGASLSPSALPLFSPSVLTDVSARSPSSPS PSLGSYGRPRSGQLSPGLSRSPSTAREGPEDTVSLPFSREVQAPSDGAHGDARITPDRAR HRLAGDEEARTDRGISDRRDEDRYWSGSRGDREVAWIPIARLPASEGTGVVVSSHPSGRF FSLSSGIVSRYFVSGRSGGDRGGHGQAPQTPRAHAALPRVGKKSPGGEGASNPSVCAFCA ALRERPGAQAELQDRAERRNDARRRGENGEDGKRRRGSEGNRKRNASVETGAEGQEARRH CMFVTGVFRFGSTTHAASRFALFLRHCCGFLSARLASPRSRFVTAWICSRPPFTTVTPCG FHSRVPFGLLCLGPVLPCLRFHGFTSLFVPLVSCFVARLLWIFLGMRWRLSLSVSVLAVT ADFARGSSGAPVSDAATGELLGIAQSTSSLYCGPGSEAPHGVDQGKGKRRKLSSGLPASW PAPSASQEAAENATANQSSAAFADAPRDHAGSWTGTRRLGNSDKAEREENGELGEPVACG YEGEGRRQRDGGRGRHGARQDAETREKKRHEGRRSGQAGCESRTLQMVVKTCIPSLYIFD LLEDS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_046090 | 70 S | SSSLSPSAA | 0.996 | unsp | NCLIV_046090 | 70 S | SSSLSPSAA | 0.996 | unsp | NCLIV_046090 | 70 S | SSSLSPSAA | 0.996 | unsp | NCLIV_046090 | 172 S | RRRESRIKQ | 0.997 | unsp | NCLIV_046090 | 202 T | RARDTHFED | 0.993 | unsp | NCLIV_046090 | 218 S | VLGLSPGDG | 0.996 | unsp | NCLIV_046090 | 377 S | SSGSSPSSF | 0.993 | unsp | NCLIV_046090 | 408 S | VSSSSPELP | 0.995 | unsp | NCLIV_046090 | 463 S | RGDSSQERW | 0.993 | unsp | NCLIV_046090 | 545 S | SQDTSGKMD | 0.991 | unsp | NCLIV_046090 | 576 T | DRRGTARQK | 0.994 | unsp | NCLIV_046090 | 581 S | ARQKSRHRK | 0.997 | unsp | NCLIV_046090 | 655 S | VSARSPSSP | 0.995 | unsp | NCLIV_046090 | 658 S | RSPSSPSPS | 0.994 | unsp | NCLIV_046090 | 675 S | SGQLSPGLS | 0.991 | unsp | NCLIV_046090 | 715 T | DARITPDRA | 0.99 | unsp | NCLIV_046090 | 736 S | DRGISDRRD | 0.998 | unsp | NCLIV_046090 | 746 S | DRYWSGSRG | 0.994 | unsp | NCLIV_046090 | 748 S | YWSGSRGDR | 0.991 | unsp | NCLIV_046090 | 824 S | VGKKSPGGE | 0.994 | unsp | NCLIV_046090 | 878 S | RRRGSEGNR | 0.997 | unsp | NCLIV_046090 | 887 S | KRNASVETG | 0.997 | unsp | NCLIV_046090 | 937 S | ARLASPRSR | 0.994 | unsp | NCLIV_046090 | 1112 S | DHAGSWTGT | 0.995 | unsp | NCLIV_046090 | 1122 S | RLGNSDKAE | 0.992 | unsp | NCLIV_046090 | 19 S | RRSRSLSRP | 0.991 | unsp | NCLIV_046090 | 21 S | SRSLSRPSL | 0.991 | unsp |