

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
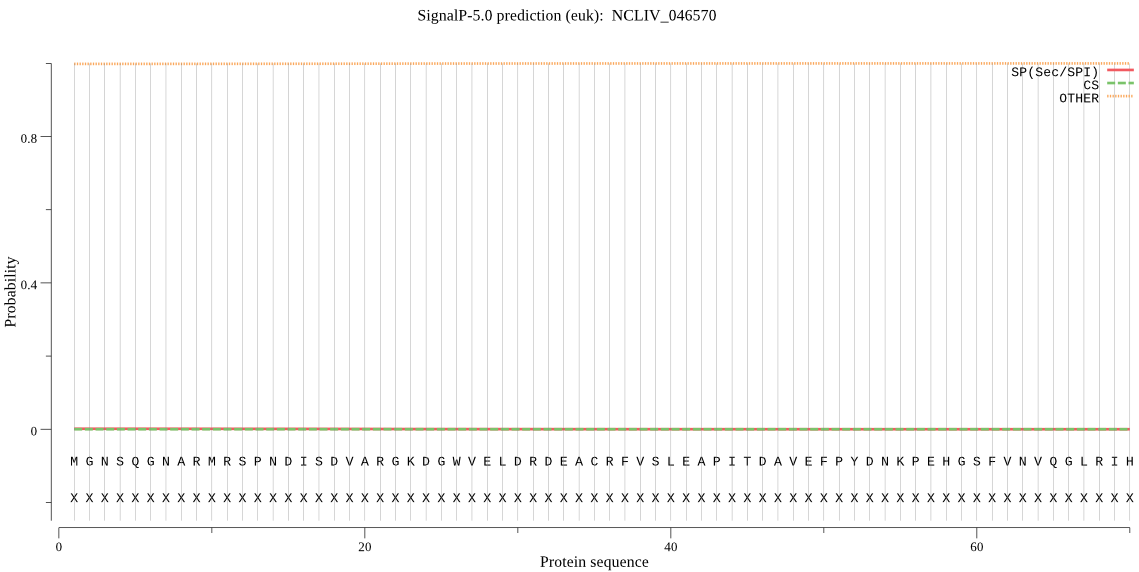
| NCLIV_046570 | OTHER | 0.999988 | 0.000011 | 0.000001 |
MGNSQGNARMRSPNDISDVARGKDGWVELDRDEACRFVSLEAPITDAVEFPYDNKPEHGS FVNVQGLRIHTYFWYPQASQSRTSGAGLRESTSRPDASASPLLPQRHSSCSPTASFSTLG AGQAAPSSSLDRNRGPTKVAHAGFYPPHPSPSVTSVPRHALPTCNGRRAYTADHLGAASG SLPAPEQALLDKATAVCGARGVVLLLHGYCNHSRLAWLSKPLPPGHRRRDPTVVGYPTVS EALSVPAAVASNSSCRSEADEQSFDFSPAATLSTAGSWKNALEGTSEFKPDPPGRVASRS RAASLTAEEPAGGCDRTEGWVETVGDASVSASWHIQYQGSWVEALNRAGFLVAAMDFQGH GLSEGWQGKRGSVNRLDDLALDVIQFITIIRRRCQFLRGVYRHCVHDGKHRSGSREVKEG AERRPDSERQTSSKPRRGMPTSEMEPNGGETKEQSTQESNGGIDNDEENPPIFLLGVSLG GWAAARTIQLLGDPKAVASLFLVCDPNGPLPQFSGILTPHAAASPPHSGASAGPLPFPIE KQLTSSLSAARVSRQRQSSSFASLGALSSVGQTQTSLTTAGVSGCVLLSPMLDLSFVKSA TKYNLYRHLMVRLAEWAPNMVVSRPLPNKEYPYLEAHMQRDKYTYKGGSRLRMVREMFEG TDTILHPECLSLISESTCGALLVMHNLLDGVCGVEGSLRLFHGATRIRDKSFLAVNAYVD GTASTQHLNPVEFSTGAPAEVLARFGTGALPAPASTLAGCGSPEVLQPLQETREGVKAAG GSMDLEGQAAAQDPHATPCAVTAEIRCGLAHDLSEGTSESVERKRTPSQHRHQGTEARDS VSGPAVGKEEVGTEEVCDVTSLNAHEGGKSQQQENVERAVPFQATGREFSLVFF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_046570 | 91 S | GLRESTSRP | 0.995 | unsp | NCLIV_046570 | 91 S | GLRESTSRP | 0.995 | unsp | NCLIV_046570 | 91 S | GLRESTSRP | 0.995 | unsp | NCLIV_046570 | 92 T | LRESTSRPD | 0.993 | unsp | NCLIV_046570 | 304 S | SRAASLTAE | 0.995 | unsp | NCLIV_046570 | 414 S | HRSGSREVK | 0.995 | unsp | NCLIV_046570 | 427 S | RRPDSERQT | 0.997 | unsp | NCLIV_046570 | 432 S | ERQTSSKPR | 0.997 | unsp | NCLIV_046570 | 559 S | QRQSSSFAS | 0.991 | unsp | NCLIV_046570 | 828 S | KRTPSQHRH | 0.995 | unsp | NCLIV_046570 | 840 S | EARDSVSGP | 0.997 | unsp | NCLIV_046570 | 12 S | ARMRSPNDI | 0.994 | unsp | NCLIV_046570 | 84 S | QSRTSGAGL | 0.996 | unsp |