

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
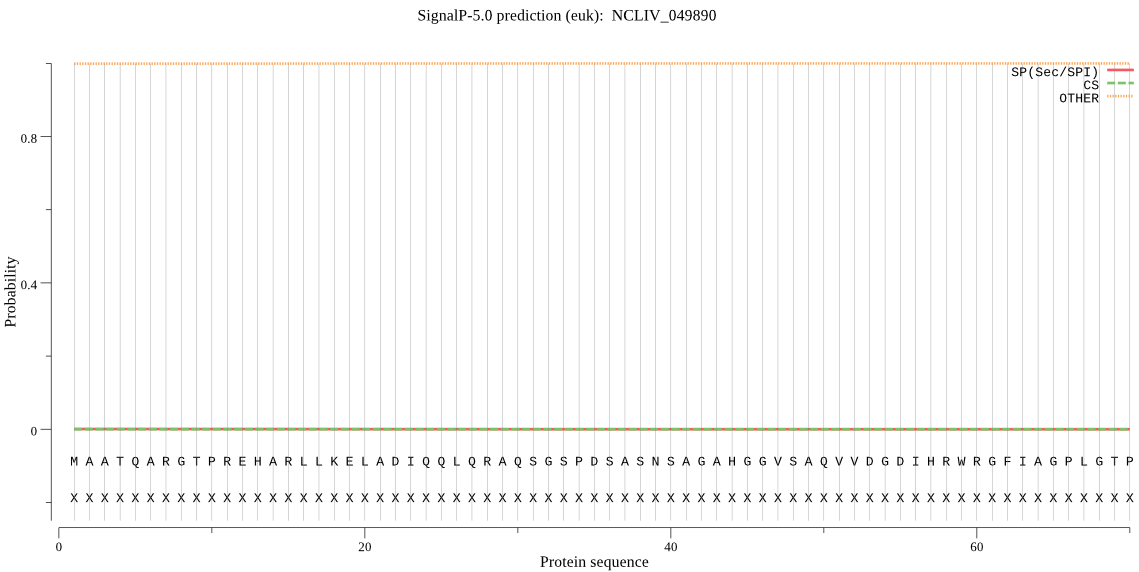
| NCLIV_049890 | OTHER | 0.994167 | 0.000160 | 0.005674 |
MAATQARGTPREHARLLKELADIQQLQRAQSGSPDSASNSAGAHGGVSAQVVDGDIHRWR GFIAGPLGTPYEGGHFTLDIVIPPDYPYNPPKMKFVTKIWHPNISSQTGAICLDILKHEW SPALTIRTALLSIQAMLADPVPTDPQDAEVAKMMMENHPLFVKTAKLWTETFAKEAQDSQ EDKVRKLTEMGFAEDQAREALRKHDWDETMALNSLVEGSDNTEKDTGKLAKEHRREAEKR RAQIPPAQLKNLGLSTVKRISKTRRSWFLILSSMIHSSALEISQTYSKCQGAA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_049890 | 261 S | VKRISKTRR | 0.996 | unsp | NCLIV_049890 | 261 S | VKRISKTRR | 0.996 | unsp | NCLIV_049890 | 261 S | VKRISKTRR | 0.996 | unsp | NCLIV_049890 | 9 T | QARGTPREH | 0.996 | unsp | NCLIV_049890 | 179 S | EAQDSQEDK | 0.998 | unsp |