

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_051530 | mTP | 0.088741 | 0.443905 | 0.467353 | CS pos: 66-67. RCF-AS. Pr: 0.6535 |
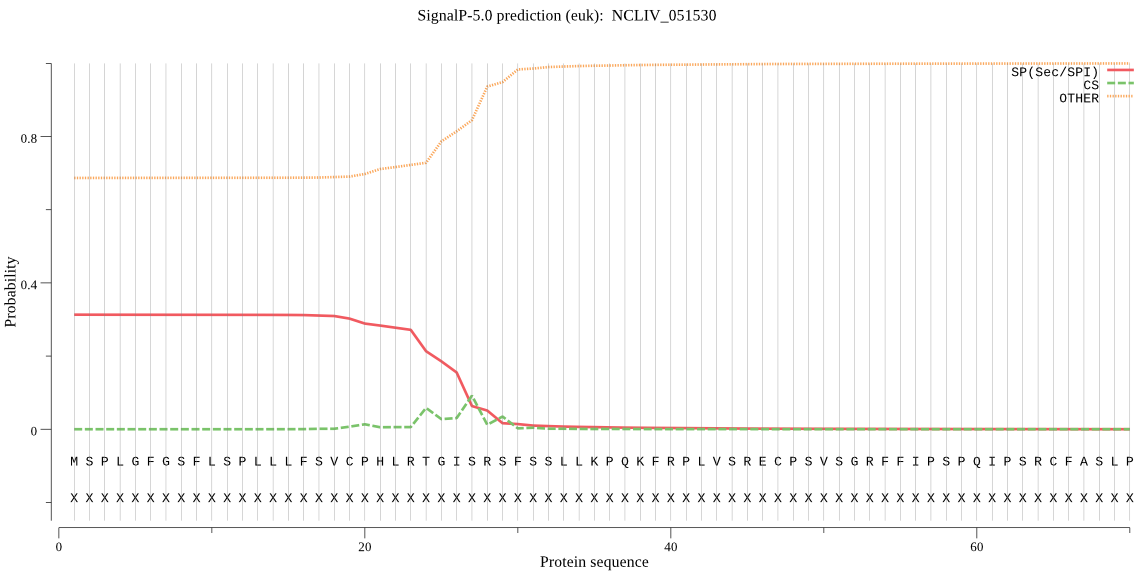
MSPLGFGSFLSPLLLFSVCPHLRTGISRSFSSLLKPQKFRPLVSRECPSVSGRFFIPSPQ IPSRCFASLPASSKMAIKVLVPVAHDSEEIEAVCIVDTLRRAGAEVMVASVEDNKLVRMS RGVCVQADKLISEVQNEVFDCIAVPGGMPGAERCRDSAILTKMLKAHKEQGKFIAAICAS PAVVFQTHGLLEGEKAVAYPCFMDKFPANVRGEGRVCVSNKIVTSVGPSSAIEFALKLVE VLYNEEQAKKIASQLLCS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_051530 | 110 S | VMVASVEDN | 0.996 | unsp |