

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_051630 | OTHER | 0.959727 | 0.000857 | 0.039415 |
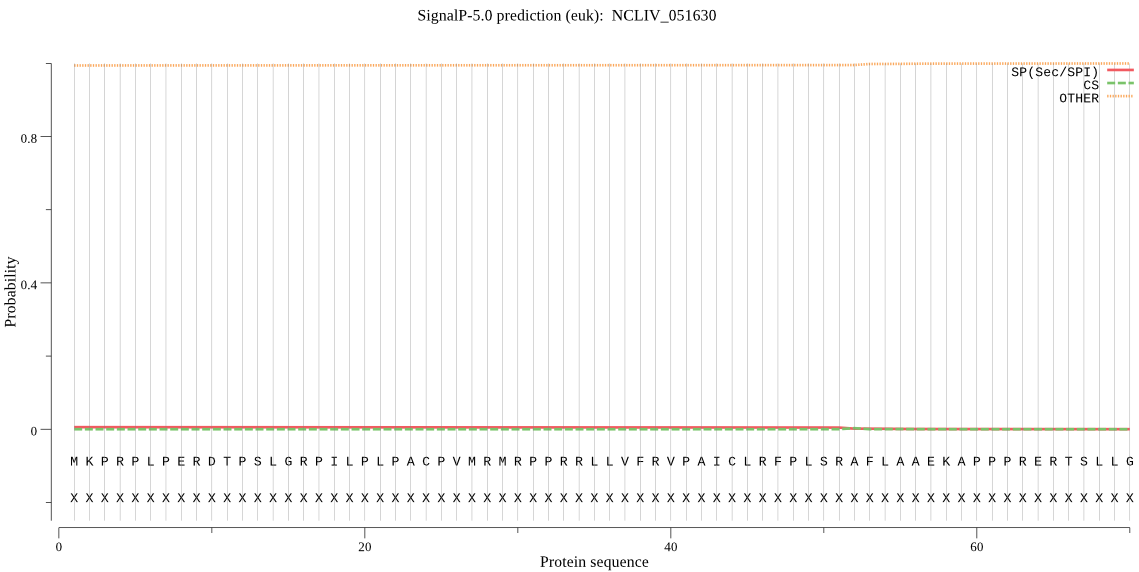
MKPRPLPERDTPSLGRPILPLPACPVMRMRPPRRLLVFRVPAICLRFPLSRAFLAAEKAP PPRERTSLLGLASSRVTAASPLRTSLRGVYVHLALEQGDLLRRPLSPQRTRFVSAPLRST NGGRNTNAAPPRRRLSVLSSPVLQTPFSLQPLPKQRTYSERAAIGPKSDMRRKALTSSFS MRRNARQTARISPSVALTPSLGKQAESGREWSQMDTATACWHATASANKAKAKKGGSGRE EKTAQKTEEPESEKTHAKAENGFSDAMLSSLYWKRKTDALDTRSVLDLTPRCLDDYLPAV ASPVAGRSGDGARRGISAPSSLVEPFGADAASLPHAKAESDKTSRQTVQEIQARAATLAQ AARKPLEVYRSYAASLQEKLELLRERERRAKSLLETLPARGERLLSALKVPPPPPLPLVP VRWNRESPMRCDAEALAAAQTLLTCSDPSSVLIDKFNIGLAGGQLECLYGSNWLNDEVIN FYMQMLQERNEKQRALGQNIWKTFFFNTFFYAKLTGGHSADVTYDYASVRRWTRRQNVDI FAVDLVLIPLHVNRLHWTLGVVDMRKGKRKIYFFDSLGGKNKTWFLTMRRYLQDEHTDKH EKPLEDIDEWCIPEDFASEKYTPQQANGFDCGVFICQMAECIADGRSFDFSQKDIPRIRH KMAPVPASLCVSLSLLLFPFVNPVLSFRSAAHC
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_051630 | 136 S | RRRLSVLSS | 0.996 | unsp | NCLIV_051630 | 136 S | RRRLSVLSS | 0.996 | unsp | NCLIV_051630 | 136 S | RRRLSVLSS | 0.996 | unsp | NCLIV_051630 | 159 S | QRTYSERAA | 0.994 | unsp | NCLIV_051630 | 207 S | KQAESGREW | 0.995 | unsp | NCLIV_051630 | 212 S | GREWSQMDT | 0.994 | unsp | NCLIV_051630 | 237 S | KKGGSGREE | 0.997 | unsp | NCLIV_051630 | 252 S | EEPESEKTH | 0.996 | unsp | NCLIV_051630 | 321 S | SAPSSLVEP | 0.992 | unsp | NCLIV_051630 | 392 S | RRAKSLLET | 0.996 | unsp | NCLIV_051630 | 427 S | WNRESPMRC | 0.993 | unsp | NCLIV_051630 | 528 S | YDYASVRRW | 0.992 | unsp | NCLIV_051630 | 651 S | SFDFSQKDI | 0.998 | unsp | NCLIV_051630 | 80 S | VTAASPLRT | 0.992 | unsp | NCLIV_051630 | 106 S | RRPLSPQRT | 0.997 | unsp |