

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_057550 | SP | 0.005343 | 0.994483 | 0.000174 | CS pos: 29-30. SLA-DS. Pr: 0.7701 |
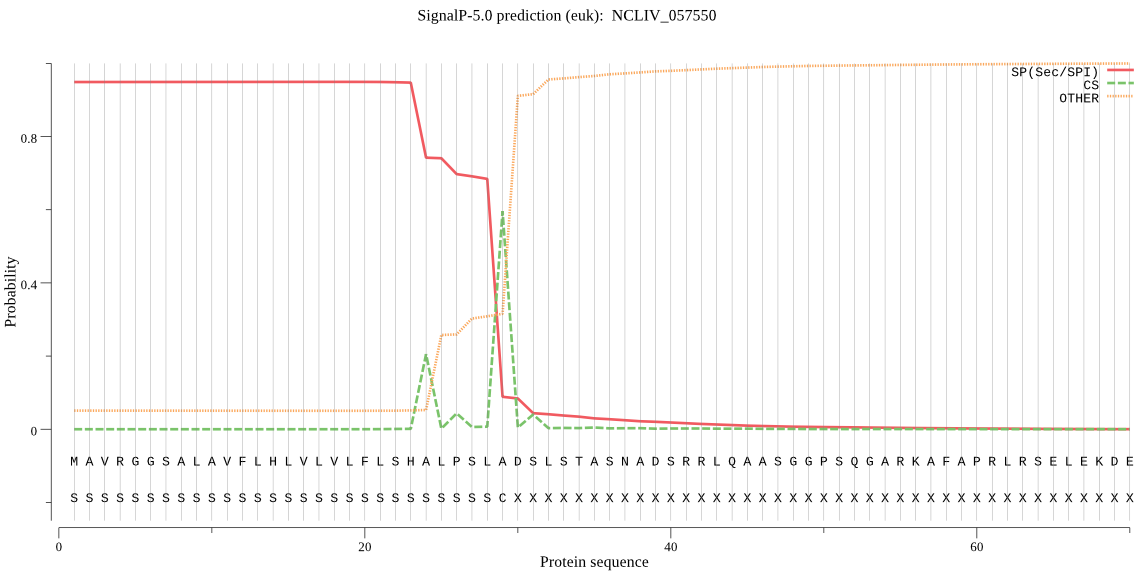
MAVRGGSALAVFLHLVLVLFLSHALPSLADSLSTASNADSRRLQAASGGPSQGARKAFAP RLRSELEKDEGAFGAVEAEECTAGTGKKCRSREWAPVTIVDDGLFDVINPTISGDEKDEL NWRVHQQEEGAGTPESNGERSEPRDQAKARTLTKGGDAGRKPRNDQDKTKPRVDASIDGK LAGPDDGDSSPIGASSATPAEKPEDAVPKPGRADWNPWKLLDENPLPNNRENWEEEEDTE DADIDPDVLPDAPDGSPRDVIPDPGDDVAPDPGDDVIPDPGDDVIPDPGDDVIPDPGDDV IPDPGDDVAPDPGDDVIPDPGDDVIPDPGDDVAPDPGDDVIPDPGDDVAPDPGDDVAPDP GDDVIPDPGDDVIPDPGGDVAPDPGDDVIPDPGDDVIPDPGDDVAPDPGDDVIPDPDGEA VHPPGKEKSPGDGSGGRTRRGSSPSEPTKKEPDTSSSTPSPGDESTPVDSGVTKWSGAGV YRGTAITLNAQWIDTHAVGQQGIRLEYQESVFPAPPVLRKVAVKSTWNQTVKIFVRTTRA RFSPAPRPIFTPPETATFSDSSLSPWWSTWTSSAVHAAMAATSSVAGFLFGTDAGWSSEE KPEAKGPTITSLKNMLKQYRRFKRKHPGKPGELVIDFCKDEYDSSSVSHAASSPSVPESS STGPVRASHKSSKDSPSSSAPSPSASSSFVGQNTRRDNKAETEGVRRTVSSSESRSAQEE NQNRKKRPKKRRKALANAHNILETQDGLALVNRIRDKMTELFGTPSTAVQALVSTGHVIV TLPAEGVDRSRLQALLETIQDTFGNNIEQWSFSEIVQLRTVVRESDPRPHAQMVAKMESL SGKSAQSSKASSLSARSEIARKKSPFEGSMPTDSALESKLYGMYMITATNAWIHGESGDG TVVAVVDSGISPHPDLDCNFWKNPYEEQDGRDDDGNGLIDDQRGYDFEENRSEPDDRNGH GTHVAGIIGACANGGGTVGGAPKTQLMALRFIDKNGLGSMANSIRALNYAIDHGVQVINN SWGGPQSTAVLRRLVHMSAAARNGKGILLLNAAGNESSNNDVYPSYPANWDQDNTISVAA IDINGELADFSNYGREAVDMGAPGVKIYSTKPGGDYQHMSGTSMATPHVSAVAALIFGAF LRNGYDAPAAEVKDIIRLTASPVDQLKDVTRWGTRPNAGDAVLMARMGGMFAQAKCEDMV FELDAGSAHIVDIHLMGYRRGTYTADLTFDVFSENGYRLDQIRVPMKLVISSEPREDDND SRAAAAFSDVVSTFSAHPSEGFEELCRLQAKIHAERKASPAMIIAICVGVLAVVLALLLA ILCCRSG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_057550 | 429 S | GKEKSPGDG | 0.997 | unsp | NCLIV_057550 | 429 S | GKEKSPGDG | 0.997 | unsp | NCLIV_057550 | 429 S | GKEKSPGDG | 0.997 | unsp | NCLIV_057550 | 442 S | TRRGSSPSE | 0.997 | unsp | NCLIV_057550 | 443 S | RRGSSPSEP | 0.998 | unsp | NCLIV_057550 | 460 S | SSTPSPGDE | 0.997 | unsp | NCLIV_057550 | 543 S | RARFSPAPR | 0.993 | unsp | NCLIV_057550 | 597 S | DAGWSSEEK | 0.994 | unsp | NCLIV_057550 | 668 S | PVRASHKSS | 0.997 | unsp | NCLIV_057550 | 671 S | ASHKSSKDS | 0.998 | unsp | NCLIV_057550 | 675 S | SSKDSPSSS | 0.997 | unsp | NCLIV_057550 | 682 S | SSAPSPSAS | 0.996 | unsp | NCLIV_057550 | 710 S | RRTVSSSES | 0.996 | unsp | NCLIV_057550 | 712 S | TVSSSESRS | 0.994 | unsp | NCLIV_057550 | 825 S | VVRESDPRP | 0.995 | unsp | NCLIV_057550 | 847 S | KSAQSSKAS | 0.994 | unsp | NCLIV_057550 | 864 S | ARKKSPFEG | 0.998 | unsp | NCLIV_057550 | 952 S | EENRSEPDD | 0.995 | unsp | NCLIV_057550 | 1161 S | RLTASPVDQ | 0.995 | unsp | NCLIV_057550 | 91 S | KKCRSREWA | 0.993 | unsp | NCLIV_057550 | 256 S | APDGSPRDV | 0.995 | unsp |