

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
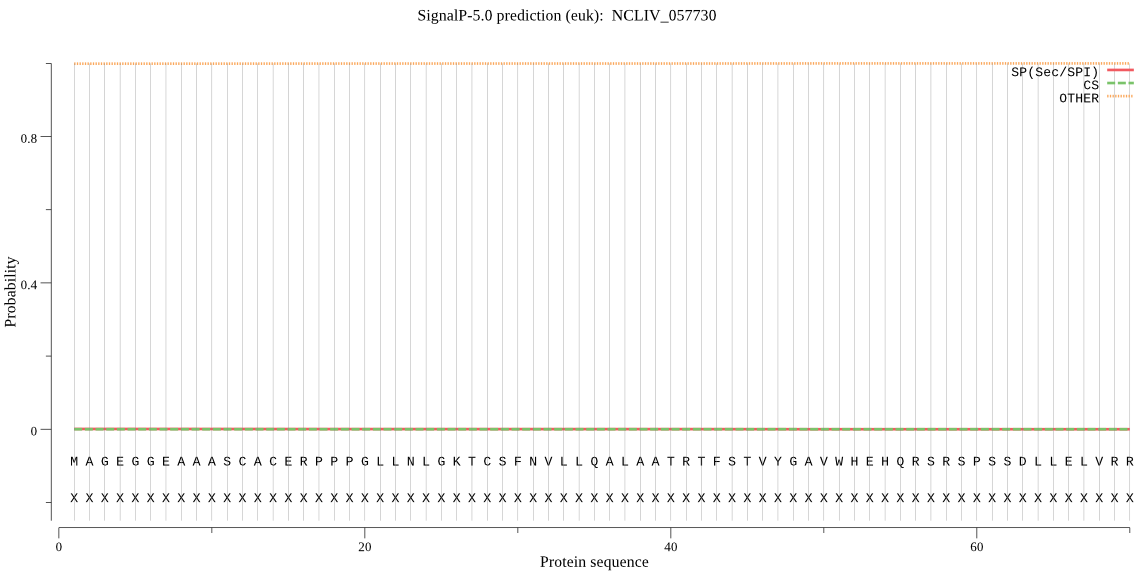
| NCLIV_057730 | OTHER | 0.999267 | 0.000241 | 0.000492 |
MAGEGGEAAASCACERPPPGLLNLGKTCSFNVLLQALAATRTFSTVYGAVWHEHQRSRSP SSDLLELVRRQRALLRRPAEAETRERGRTATSRSDGLRSSVSADASARGRPRRWRREAWC GDAAAVGRAAAPIITASRTPLNYCMADILGRMHAGEDSGASPGAAGGGASLLDNASPATA SQSPLACTQPASVDDKEDGGAPQDDPEKGQKEMMRRQLRLKRQLQTKRWWIKQEQNVLSP FDFVSQMRMLNVDFEQDVELDVEEVWRFLIQQIFTELQSEQASLASPDDVSVSSVRGLSA PTSTRREEDVGSVKGEGTPVSASVCVPARDRPGFFGGDSFPNGILRPVSGFAETGREAGR GCLANEPDGRPSRSDCSSDLTKTHQASGTVASSAQVPRPISSPQRALGPLRSSALSHSMR GGGASDPRPRHAAGAVAVPGGAFSASCTRLRTHCVRGLSPRSVPRESEVGREESERERAE RRAREKFLSASYHFLAAEAVTPRGKGAGAEENAGPWSPLRRSETTEPSAERSCSASRLAS ALPLLEGKSPGPSPRRDGDAAARCCDAGQASRVAQQSPGRSRAVPGHLKARLRLPSPPGA KLEAPVSWSDCKPAEIRTFVSSPVSGIQRAGLCGSRGRGEESVSGAPSGNADAKQEPNVK GARDAGSLAAVAQQQGATLLLRQLQQALVGRLVQVPVPCSCCCRCEAGAPASVRPRLQTQ SPGACRSRSASAPPTDRSGRVGSPETVPGGTVLSSNRERKHGSNDASPGPAASSGSPARD DRSEARSLSPRRGLLPVESRRNGVDRRGNVCASPAASPSQSCVHAASGRSSPLSVPVSSH TEAKSASLSRRSQKENLAPSALAAQDSPRASRASSPEGVNSARSGRSKGSGEAKSSSAVS AFLTPSLFPSSLATHPVCTPLAPAVATSGVQAVGLEQDDGTSGIEGLSSRGDAMLSLLSV ESFCGVLPVRLPERSRWVSSRARTNTVGLSGSSLSSTGLDASSPLLARADSTPQSADSAE TMGGSRRISPLQPITLEECLALAFQQSCGGASPSGGAAAAAEAGERKRGREAARRARPAG AFGGRRRLSRTQRRVKAGDAEKKTWRGTGPANPGSEGGKHKCLACGKWRVRRRGCAPEAA PSAGEGENATHAAGSRQGRVSGAVEVEGAERRTLSRSGTRTSTRDTTPAHSEETAKNGDR EAAPGKESQVWRDSRDAFSCYQRLVCHRCNQVHMRTFLERLPEILLFQLPRSSITPPGTF GSGAVKAAAAAAGGGFKIKAHVEFPQRLESVIAPHCVASPAFEAAWGRLMDATGRPGGSD SSGSLESDGGTDVSGRRKRRKLSSQRAQNRAGSKTAGTRGEGKAVHRRVANSVSRDERPG TGAATLSTLAGTAAAPDSIRHTLQHEAGVYTLRAVIEHQGRSGVGGHYVCYRRGAPCDLV VETRENGEHLEGSPASLTSLSQAGAVSFSAAPQGEARRGVWDDSAAVLENSPREETPEEG TWWEVNDAEVTQTTWDHVRLSQAYILVYERCDENVCCSRFGGDRRVACRAQVVSSPAFPR TGAETDGPTPAARGRSGPRCGALSGAEKNVPLVSVQSRTRRLLRAGEPCRTSGSCRRAEG RNEACRPETREPGETGVASERPQAPRQKKKTSTRDTAGEGGHEKSARGVCAGRRPRSRRG RSSESHSSVARGPRRAGEHGEDAGECRGRCIVSGEGERPVPLEPPANATERRRGRRRDDG AFLWTGESGKNVGSGNRQGRRATRQRSVASATIAADA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_057730 | 303 S | SAPTSTRRE | 0.994 | unsp | NCLIV_057730 | 303 S | SAPTSTRRE | 0.994 | unsp | NCLIV_057730 | 303 S | SAPTSTRRE | 0.994 | unsp | NCLIV_057730 | 372 S | DGRPSRSDC | 0.997 | unsp | NCLIV_057730 | 467 S | VPRESEVGR | 0.991 | unsp | NCLIV_057730 | 474 S | GREESERER | 0.992 | unsp | NCLIV_057730 | 553 S | SPGPSPRRD | 0.996 | unsp | NCLIV_057730 | 577 S | VAQQSPGRS | 0.995 | unsp | NCLIV_057730 | 596 S | LRLPSPPGA | 0.993 | unsp | NCLIV_057730 | 731 S | SRSASAPPT | 0.995 | unsp | NCLIV_057730 | 789 S | ARSLSPRRG | 0.997 | unsp | NCLIV_057730 | 817 S | SPAASPSQS | 0.99 | unsp | NCLIV_057730 | 839 S | VPVSSHTEA | 0.994 | unsp | NCLIV_057730 | 849 S | SASLSRRSQ | 0.994 | unsp | NCLIV_057730 | 852 S | LSRRSQKEN | 0.998 | unsp | NCLIV_057730 | 871 S | SPRASRASS | 0.991 | unsp | NCLIV_057730 | 874 S | ASRASSPEG | 0.997 | unsp | NCLIV_057730 | 875 S | SRASSPEGV | 0.996 | unsp | NCLIV_057730 | 949 S | EGLSSRGDA | 0.993 | unsp | NCLIV_057730 | 1029 S | SRRISPLQP | 0.997 | unsp | NCLIV_057730 | 1089 S | RRRLSRTQR | 0.996 | unsp | NCLIV_057730 | 1142 S | EAAPSAGEG | 0.993 | unsp | NCLIV_057730 | 1175 S | RRTLSRSGT | 0.995 | unsp | NCLIV_057730 | 1177 S | TLSRSGTRT | 0.993 | unsp | NCLIV_057730 | 1182 S | GTRTSTRDT | 0.998 | unsp | NCLIV_057730 | 1191 S | TPAHSEETA | 0.997 | unsp | NCLIV_057730 | 1372 S | RVANSVSRD | 0.995 | unsp | NCLIV_057730 | 1491 S | VLENSPREE | 0.996 | unsp | NCLIV_057730 | 1496 T | PREETPEEG | 0.995 | unsp | NCLIV_057730 | 1652 S | KKKTSTRDT | 0.998 | unsp | NCLIV_057730 | 1677 S | RRPRSRRGR | 0.995 | unsp | NCLIV_057730 | 1682 S | RRGRSSESH | 0.997 | unsp | NCLIV_057730 | 59 S | QRSRSPSSD | 0.995 | unsp | NCLIV_057730 | 1683 S | RGRSSESHS | 0.996 | unsp | NCLIV_057730 | 92 S | RTATSRSDG | 0.996 | unsp |