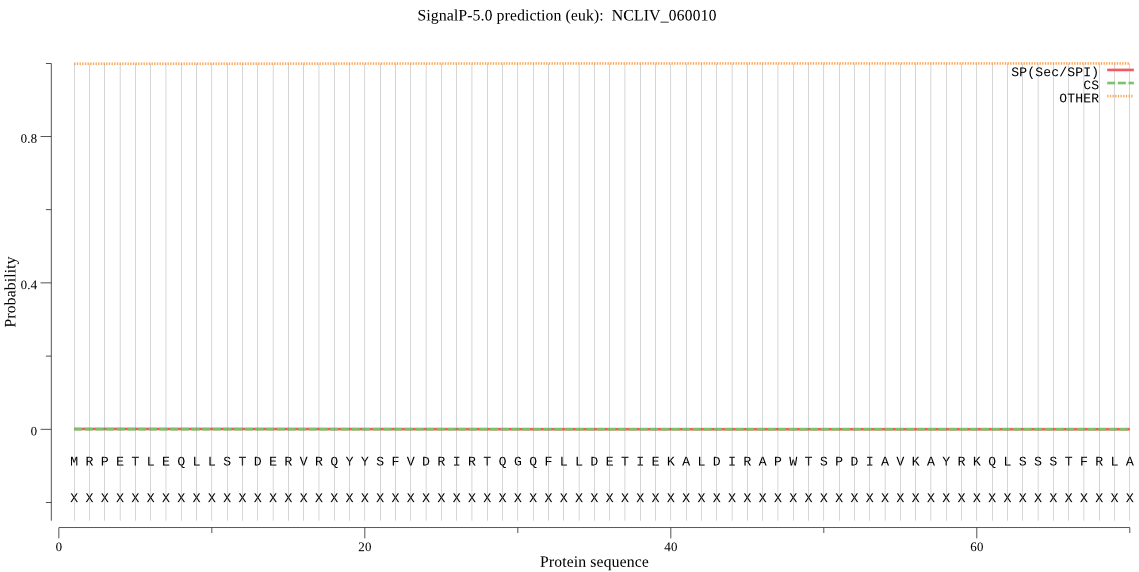
Fasta :-
MRPETLEQLLSTDERVRQYYSFVDRIRTQGQFLLDETIEKALDIRAPWTSPDIAVKAYRK
QLSSSTFRLAGQPDVFSYTQIRELMRSENQEIRHHGQQAIKDGIVENRIDKFASLALNMV
VGSWSIETRERGYPSPRSRRNVLNRISDEIVDALIAAVKKEGVELTRRALLLKKQLLTAT
GKIKAFSFVDRLAPLTLSRDSSMDANARIPWCQAVEKVKKGYASFSPVFVRLFENLLKEG
RIDAAPAEGKQTGAFCSGGTPATGPFILMTHTGVKANVQTLAHEAGHAIHFLLAYEQGNL
QYHPPLPLAETASLMGETIVFYHLLDQTDSPQKVLEMLIEHVDSLVGTITRQVSFDRFEE
LVHEARLSGIVGDEDFDQMWLQSTREFFGDEGDVFDDYAGIEKDWSTVHHFTKAPFYVYS
YALADLLVGSLLAARSKNNGSDFEEKFISFLQAGDTADLETALRPFGLDPKTPSFWREAI
GMRLGTAVERAERIAAELGYIEATGY