

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_061740 | SP | 0.014959 | 0.983265 | 0.001776 | CS pos: 20-21. ALA-IL. Pr: 0.2600 |
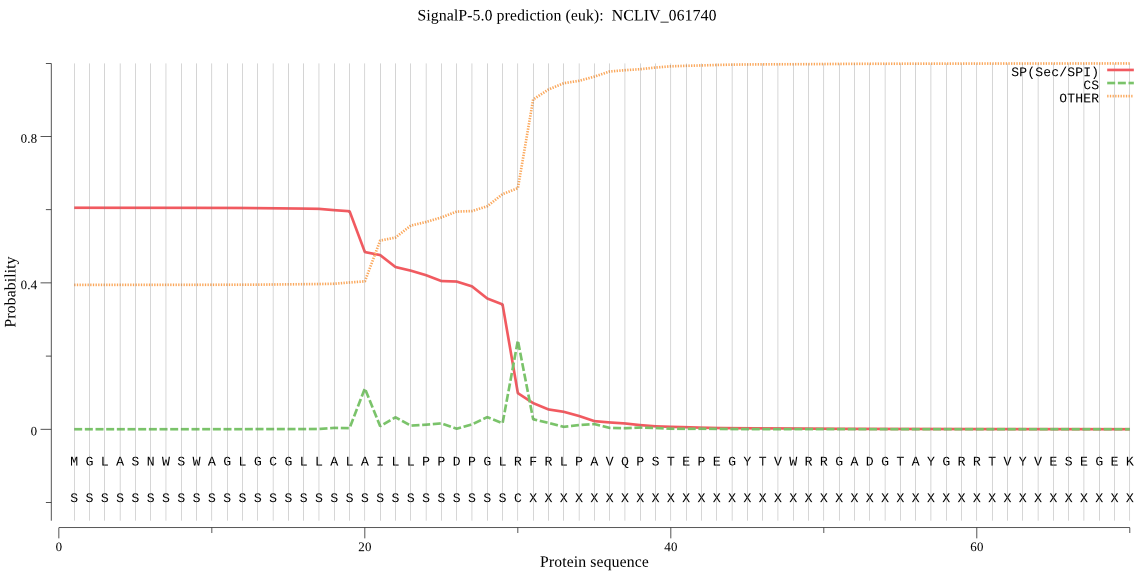
MGLASNWSWAGLGCGLLALAILLPPDPGLRFRLPAVQPSTEPEGYTVWRRGADGTAYGRR TVYVESEGEKLHAWLYLPPNSRQNAPLYMVCHGFGAVMAVADVAFAEKIQESGIAAIAFD YRTWGFSGGAPRQVVDPHMQLQDMRAVLQHVVDTDGFQGTVDPTNIHLFGTSYAGGHVLV TAAQLASENSPLLRRVRTVTSVVPLIDGKEQTKKALQQRSFFRTLRYAAAILADLLRQAV GSHLQPIYLQVAGPRGVSSLSALELQGGELEIWRNRVLDSSRTASWTNALAARSLFHTSQ YRPIRYLEHIQTPTLLVEAQHDNTCIPELAREALEIINSRGGTNSEDRAGGKGSRDSRLA ELYRMPVNHFEIYVAPHLGRLINTTVEFAKRHGARQAASGAEPA
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NCLIV_061740 | 285 S | SRTASWTNA | 0.994 | unsp |