

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
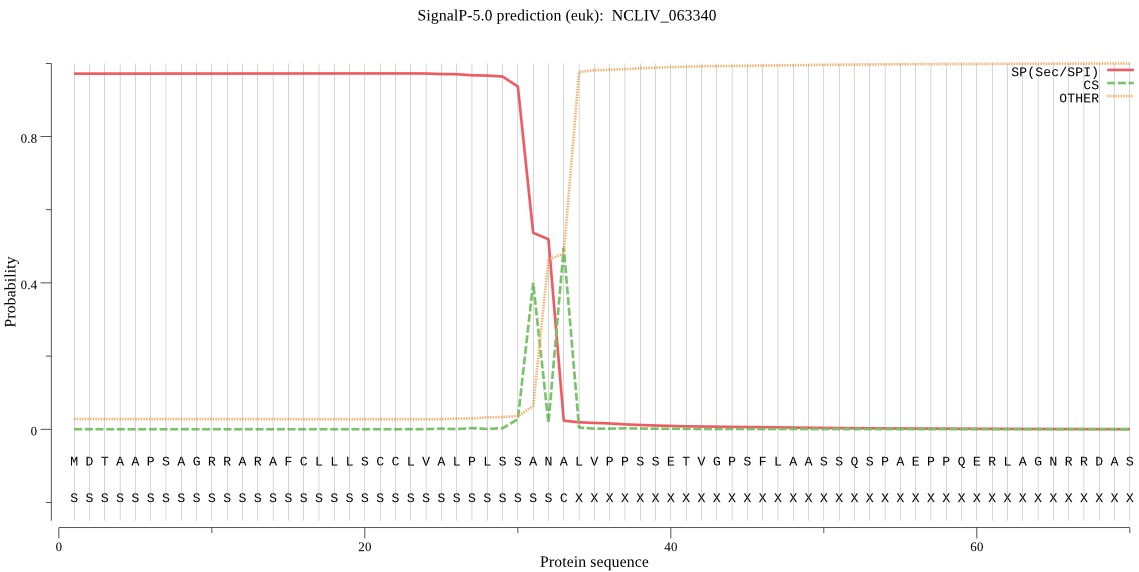
| NCLIV_063340 | SP | 0.002364 | 0.997634 | 0.000002 | CS pos: 31-32. SSA-NA. Pr: 0.6591 |
MDTAAPSAGRRARAFCLLLSCCLVALPLSSANALVPPSSETVGPSFLAASSQSPAEPPQE RLAGNRRDASHSSSLPSGTGSALRIEEIASSGSSPEVRDEAPRAPATRTTEKFIRPYHTG PSHRSSAYSATSGIPLCHTCSTFDAAKGGCRLCLHGEVASDFMMPMTPIRNIDTHREETL ARLEANHRTHLNDNKNFYVLRGKGPFGSLGNLPGTPGLDAAVAFGVVPPELPSASFAQTH SGQNSNPGDVQVGASAGGADVSNEATTSDLDLKLAETSVPILQMKDSQYVGVIGIGTPPQ FVKPIFDTGSTNLWVVGSKCKDDTCTKVTQFDPSASSTFRTASPPVHLDITFGTGRIEGS TGIDDFTVGPFLVKGQSFGLVESEGGHNMHGNIFKSINFEGIVGLAFPEMSSTGDVPIYD NIIHQGTLKENEFAFYMAKGSQVSALFFGGVDPRFYEPPIHMFPVTREHYWETPLDAIYI GDKKFCCDEGTNNYVILDSGTSFNTMPSGELGKLLEMIPPQECDLEDPEFTKDFPTITYV IGGVKFPLTPEQYLVRSKGNECKPAYMQIDVPSQFGHAYILGSVAFMRHYYTVFRRSDGT RPSLVGIARAVHSDDNRAYLSNVLNEYPGMHMRKQDLMMERSMNAPSMREL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_063340 | 126 S | SHRSSAYSA | 0.994 | unsp | NCLIV_063340 | 126 S | SHRSSAYSA | 0.994 | unsp | NCLIV_063340 | 126 S | SHRSSAYSA | 0.994 | unsp | NCLIV_063340 | 174 T | RNIDTHREE | 0.991 | unsp | NCLIV_063340 | 53 S | ASSQSPAEP | 0.992 | unsp | NCLIV_063340 | 94 S | SSGSSPEVR | 0.995 | unsp |