

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NCLIV_063390 | OTHER | 0.817887 | 0.040544 | 0.141569 |
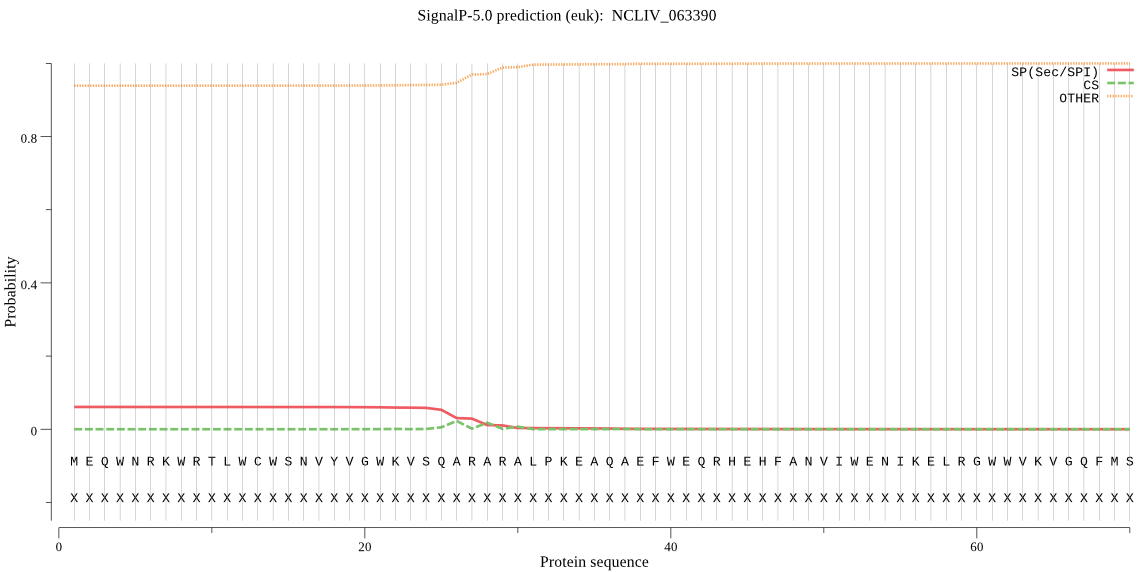
MEQWNRKWRTLWCWSNVYVGWKVSQARARALPKEAQAEFWEQRHEHFANVIWENIKELRG WWVKVGQFMSTRSDLLPQQYIHHLVKLQDMMPTSDFASIRKTIADELGDVDAIFEKIDPN ALASASIGQVHRAWLKDGSPVVVKVQHADVETLLSHDMQNLKQLSWAFGLLESGLNFAPI LEEWQKAAAKELDFRYELVHQLRAYEGLRKSGIDVKIPKPYPEFTAKKVMVMEFVNGFKI TDTEKLDAYKVDRRELMFKLCDSFAYQIHIDGLFNGDPHPGNILVEVNEATGEATPIILD WGLVKEFDSAGQLAFSKLVYAVASMNVMGLMEAFEDMGFKFKEGAGAVIDPEVYMDALRI ALRDGEVEKEETEALKQSAGQTLGAAQKAGLNRKKLQEKNPLEDWPRDIIFFVRVASLLH GLCVQLNVHLPFLQIMVKRAQECLFERYVPPSPLVYVKLGRPSRPRTQLEGRVDRLLRSL FQRGLLLGCQVAVVKDGALLVDTCIGKMGPVDARPVTSDSLFCGYCVSKGLLTTALLKLV SDGEVHLDDLVSNWWDGFIRYGKKNVTIRQLLSHRAGLHRALPANLTLSKLTDYAAMVGV IEDAPPATVPGLVGRYAYLTYGWAVSELVASVACSPVEDYIREKMLQPYGLENSIFLPLP EDGLLAERPEPATETEGREAAPKDATPPTASEPGAAAPSAEDTRDTKPTEQAAGTPSAVS AVSAVSAFSSFTKSLASGQQLLTERLGQKFLTQRREEDASSAKPASGEDAGDAGHPEPRD GSEGDAGPCSSEDAGDTAGSESPGVPGRPGACRLSREGPDVCVSAPSTPGNCGAAAAADE GRPHLARSAPPSVCGTPLGSPRTQSVASEGSRDPPEDSSAPHKTDPFPSHPVAAFSRRVS AFASSVRSFLRPKRTSSEQSGGRLASPGAETRRRGAAKSGGFPTAGLGAPDRARVRETVG SVADQQNKKRGRRLSVILHAPDAASPDSEAGSVLLYEGEPGESREASLTENLPNQAALDP KRDKAVRMYRRETSACTVDSSVASTATKAGVLEDESDRRSAYYSVSSEEPESCCCCLSPR RPSSDSRAGLVEADTSATGGCAPSQQTVSSSDNSWTETAAREEPCGYMRVNVDVEGAGRR KVSEPRGATVGPPSLGPPASHTAHSSADKEPSRSQHPPPSSASTPSSPPSSAVSSPPSSS PSPLPAHAGSGETSLKDSVESRARFPPGVCTVTAKTPLFWRITSARRRISFGNVTQQEVM EKLEAAKKYAEAQAGEKSAEEPKAASDLARRRQRLHKERAMQKLRRARNAGAGAVWRRED AERAWGGEERARETRRRRSSRRQRRDSSSGPGRGESEDRGLYPSTKDGSEESPCASLGDA FERRRSKDEEDRGREKLASRGSRLSLSGGLARRGLRSASQSKGSDCRQASLLSPASALSA LSSRGSSRRGSSSGRSAASPEDLERRGSLVSAGSPSLSGEKRAPAQRSGGSASTPLSPVS LSPGQGGDAPVHRVSALDLIRKKPHVMDPLLYDSRRIVSKRVPPTNGRFTACGLAQLYAA IAAGEVIDGPTMDAARTPATVDDSLETLLLTGGGSRVWGLGYQLYECAKVDSNQLALPPP PSPASLMRLRAAQSFTRRQSVHPSSAVAAARMLRRQFTGDREFPAGSLSPFSPGGAVSPG RGPSAEGSGDGASAHAGVGGRSPRTLSPGPGKKEERTVTGFGHGDFGGSVALCFPELKLS IAILVNDVLTGPQASREILEFLLRKFGLEPRWTTAVDFDEVLANLTPKQKRSSK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_063390 | 699 S | AAAPSAEDT | 0.997 | unsp | NCLIV_063390 | 699 S | AAAPSAEDT | 0.997 | unsp | NCLIV_063390 | 699 S | AAAPSAEDT | 0.997 | unsp | NCLIV_063390 | 766 S | AKPASGEDA | 0.998 | unsp | NCLIV_063390 | 782 S | PRDGSEGDA | 0.996 | unsp | NCLIV_063390 | 790 S | AGPCSSEDA | 0.993 | unsp | NCLIV_063390 | 815 S | ACRLSREGP | 0.996 | unsp | NCLIV_063390 | 852 S | SAPPSVCGT | 0.992 | unsp | NCLIV_063390 | 900 S | SRRVSAFAS | 0.995 | unsp | NCLIV_063390 | 916 S | PKRTSSEQS | 0.997 | unsp | NCLIV_063390 | 975 S | GRRLSVILH | 0.995 | unsp | NCLIV_063390 | 1007 S | SREASLTEN | 0.994 | unsp | NCLIV_063390 | 1060 S | SDRRSAYYS | 0.992 | unsp | NCLIV_063390 | 1066 S | YYSVSSEEP | 0.991 | unsp | NCLIV_063390 | 1083 S | PRRPSSDSR | 0.993 | unsp | NCLIV_063390 | 1084 S | RRPSSDSRA | 0.996 | unsp | NCLIV_063390 | 1109 S | QQTVSSSDN | 0.994 | unsp | NCLIV_063390 | 1114 S | SSDNSWTET | 0.997 | unsp | NCLIV_063390 | 1143 S | RRKVSEPRG | 0.993 | unsp | NCLIV_063390 | 1165 S | HTAHSSADK | 0.994 | unsp | NCLIV_063390 | 1214 S | SGETSLKDS | 0.998 | unsp | NCLIV_063390 | 1218 S | SLKDSVESR | 0.993 | unsp | NCLIV_063390 | 1278 S | AGEKSAEEP | 0.993 | unsp | NCLIV_063390 | 1339 S | RRRRSSRRQ | 0.998 | unsp | NCLIV_063390 | 1340 S | RRRSSRRQR | 0.998 | unsp | NCLIV_063390 | 1347 S | QRRDSSSGP | 0.998 | unsp | NCLIV_063390 | 1364 S | GLYPSTKDG | 0.997 | unsp | NCLIV_063390 | 1369 S | TKDGSEESP | 0.995 | unsp | NCLIV_063390 | 1386 S | ERRRSKDEE | 0.998 | unsp | NCLIV_063390 | 1405 S | GSRLSLSGG | 0.993 | unsp | NCLIV_063390 | 1446 S | SSRGSSRRG | 0.996 | unsp | NCLIV_063390 | 1447 S | SRGSSRRGS | 0.997 | unsp | NCLIV_063390 | 211 S | GLRKSGIDV | 0.996 | unsp | NCLIV_063390 | 1451 S | SRRGSSSGR | 0.996 | unsp | NCLIV_063390 | 1452 S | RRGSSSGRS | 0.995 | unsp | NCLIV_063390 | 1453 S | RGSSSGRSA | 0.993 | unsp | NCLIV_063390 | 1459 S | RSAASPEDL | 0.998 | unsp | NCLIV_063390 | 1515 S | VHRVSALDL | 0.993 | unsp | NCLIV_063390 | 1640 S | TRRQSVHPS | 0.996 | unsp | NCLIV_063390 | 1672 S | LSPFSPGGA | 0.993 | unsp | NCLIV_063390 | 1678 S | GGAVSPGRG | 0.991 | unsp | NCLIV_063390 | 1707 S | PRTLSPGPG | 0.995 | unsp | NCLIV_063390 | 1793 S | QKRSSK--- | 0.994 | unsp | NCLIV_063390 | 463 S | LGRPSRPRT | 0.99 | unsp |