

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
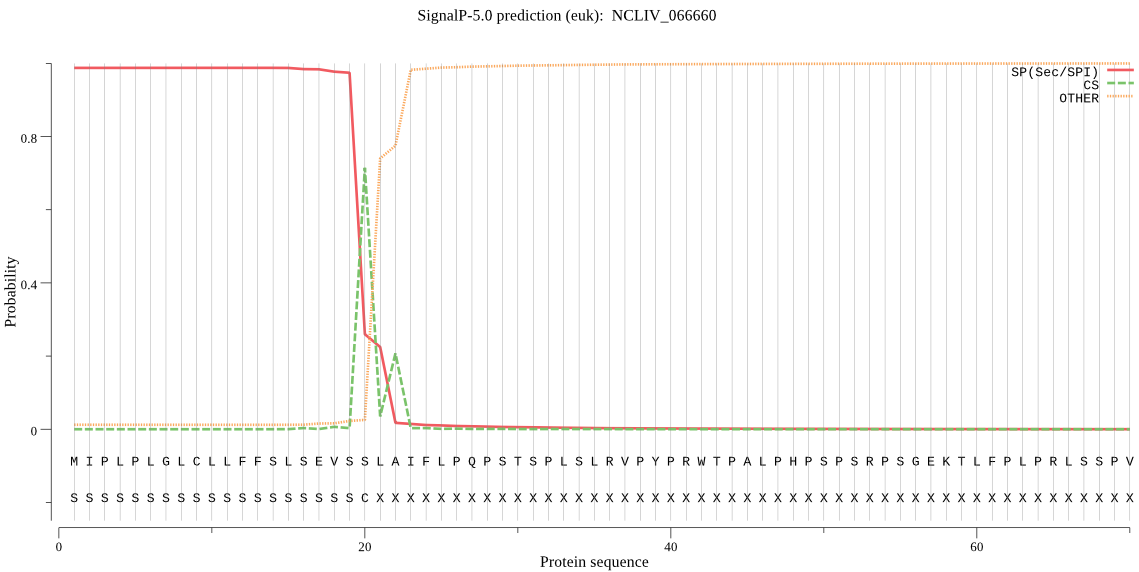
| NCLIV_066660 | SP | 0.017668 | 0.982315 | 0.000016 | CS pos: 20-21. VSS-LA. Pr: 0.6478 |
MIPLPLGLCLLFFSLSEVSSLAIFLPQPSTSPLSLRVPYPRWTPALPHPSPSRPSGEKTL FPLPRLSSPVSPFPPASALSISSVYRPPTSRVTAPQTEFPCSPQPSFSASLASPVSAGPS GVFPILVHASPVSPVAFCAFPASRKTVALARRSGAETPGRRQETAAHSLGVSQSTPAFFP SPLRLQFSRAPPPTRRRYSFSELLEHAGETSAEKASVPFPFSLNGDSRFSYTSSAFSRLA SLRIADSGNETDRGERREAAEDSQTLGEGLHPGEQLSTADEPSRRVSGESVLSPRLSQKA SVAPRNETSETARSPQLSNTVRNLEERHGDFPTSCSDSHPSSSDSPGSPDFRERLGGPRD ASSVSSPSSSRSLPSFGTRGEIRRKPPRVTSSAHSAAADRGAPFRLFSFSPPSLSLFLPL RRSSPHRSIPLFSPLSSPLFSALSSPPSSSRFSLSSRASLSSSPSPGQSSSAWYRWVHAF ASTVCSPFSKLRTCAASFLSAGGLKPAAPPSLESDASRREALGLARLRGVFRKIASRVSA LGSQANARLRDSFLAQAALVLAIYFVHFAYISQQSFVLPVQLLPNRRGLFQHVQGDSLAG WAALGALLLSRNAATRDSADCNSTPSSGPSFSSESSARSASFAATPSFSGRSSNANPSSE SPLSASGENLSPCVSTPRESSGRQGGASCDAGADHSTPAEKRNLFNAAASACFENGEEKT KCKGDRPKRLSSVSQRLPWQGPFPAFWKSATLLLALFGSYVLSGYVACGLDLLFYFLHAA GVVSLDLAMHRSLQVLLAHLAWVFMGMSLFKLSSKHFFRSAWASPPSDAHASRLHAETPD APRPGVDPPLCRGTGGDAVKLSRADFGIDRETTGESATLAGRVEAPAAGPAAQEGHSDRN EETDAASERSTEGLRVASSSSASAPGPSRPVSFLQSLPGGPRVQEGGERFLASEASSEAR LNLDGVAPGDSRSAASARVFEGCLFASDIGGEEAPRGRGIQTASVSSSGRFSRVCDAFRR LWRKEARPRALGRGQESQAHRAPRASRGQAKAEAAAPRFAGSCLDTAAGTCTDCSAAAGT RGRPMFARRAHSRWFTFRGRGKGGLWAWWVVSGYLVSCLFFNLTEFVNEALMDLLPDEPQ GPTIVQHIMNPTLNTRWSFLVGALAPCLSAPWWEELLYRGFCLPLFSQVMVLPAAATLSS LLFAVHHMNVQTVLPLWVLGLTWTAVYLQSQNLLTTVLIHAMWNSRIFLGNLFGL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_066660 | 153 S | LARRSGAET | 0.996 | unsp | NCLIV_066660 | 153 S | LARRSGAET | 0.996 | unsp | NCLIV_066660 | 153 S | LARRSGAET | 0.996 | unsp | NCLIV_066660 | 199 S | RRRYSFSEL | 0.998 | unsp | NCLIV_066660 | 230 S | DSRFSYTSS | 0.996 | unsp | NCLIV_066660 | 287 S | SRRVSGESV | 0.997 | unsp | NCLIV_066660 | 297 S | SPRLSQKAS | 0.997 | unsp | NCLIV_066660 | 334 S | DFPTSCSDS | 0.994 | unsp | NCLIV_066660 | 341 S | DSHPSSSDS | 0.996 | unsp | NCLIV_066660 | 345 S | SSSDSPGSP | 0.997 | unsp | NCLIV_066660 | 366 S | SSVSSPSSS | 0.996 | unsp | NCLIV_066660 | 395 S | SSAHSAAAD | 0.991 | unsp | NCLIV_066660 | 424 S | LRRSSPHRS | 0.998 | unsp | NCLIV_066660 | 453 S | SSRFSLSSR | 0.991 | unsp | NCLIV_066660 | 459 S | SSRASLSSS | 0.993 | unsp | NCLIV_066660 | 517 S | ESDASRREA | 0.994 | unsp | NCLIV_066660 | 676 T | PCVSTPRES | 0.993 | unsp | NCLIV_066660 | 680 S | TPRESSGRQ | 0.996 | unsp | NCLIV_066660 | 681 S | PRESSGRQG | 0.995 | unsp | NCLIV_066660 | 731 S | PKRLSSVSQ | 0.996 | unsp | NCLIV_066660 | 897 S | QEGHSDRNE | 0.996 | unsp | NCLIV_066660 | 907 S | TDAASERST | 0.995 | unsp | NCLIV_066660 | 1008 S | SVSSSGRFS | 0.991 | unsp | NCLIV_066660 | 1012 S | SGRFSRVCD | 0.991 | unsp | NCLIV_066660 | 1046 S | APRASRGQA | 0.994 | unsp | NCLIV_066660 | 1092 S | RRAHSRWFT | 0.99 | unsp | NCLIV_066660 | 55 S | PSRPSGEKT | 0.997 | unsp | NCLIV_066660 | 89 T | YRPPTSRVT | 0.994 | unsp |