

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
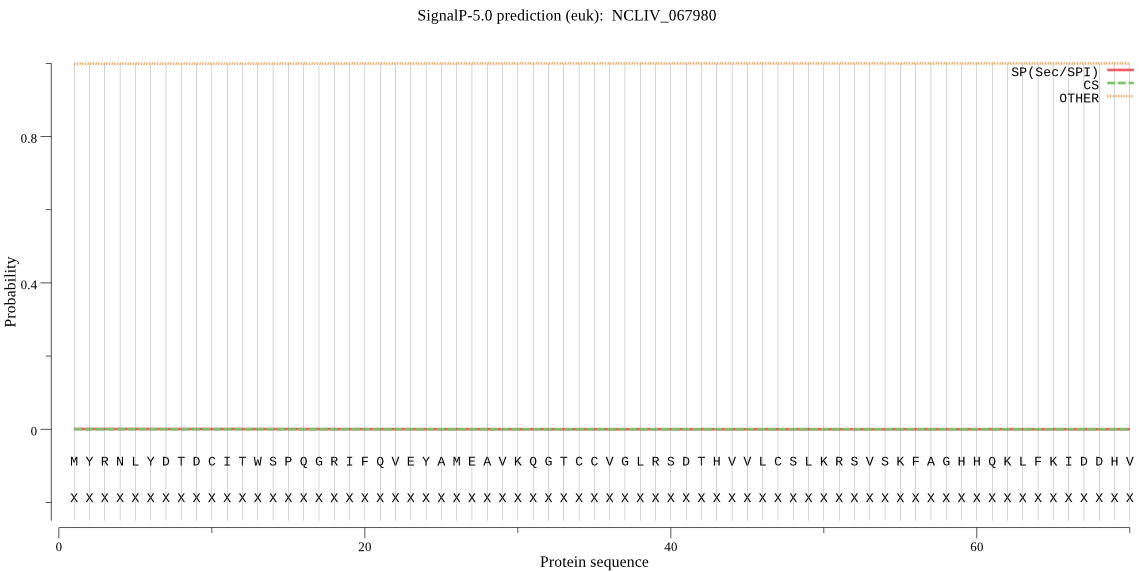
| NCLIV_067980 | OTHER | 0.999904 | 0.000062 | 0.000034 |
MYRNLYDTDCITWSPQGRIFQVEYAMEAVKQGTCCVGLRSDTHVVLCSLKRSVSKFAGHH QKLFKIDDHVGVAMSGITADAKVISNFMRNECFHHKYVYDAPIPVGRLVLMVADKSQANT QRSGKRPFGVGLLAAGFDDAGPHLFETCPSGNYFESYAMAFGARSQSSKTYLERNFESFP GLSPEELELHAMKALNASLAADAELTADTVSMAIVGKNQPWKELSLDEIQALLDKMNVEP SGEDVQM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_067980 | 241 S | NVEPSGEDV | 0.994 | unsp | NCLIV_067980 | 241 S | NVEPSGEDV | 0.994 | unsp | NCLIV_067980 | 241 S | NVEPSGEDV | 0.994 | unsp | NCLIV_067980 | 123 S | NTQRSGKRP | 0.996 | unsp | NCLIV_067980 | 183 S | FPGLSPEEL | 0.997 | unsp |