

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
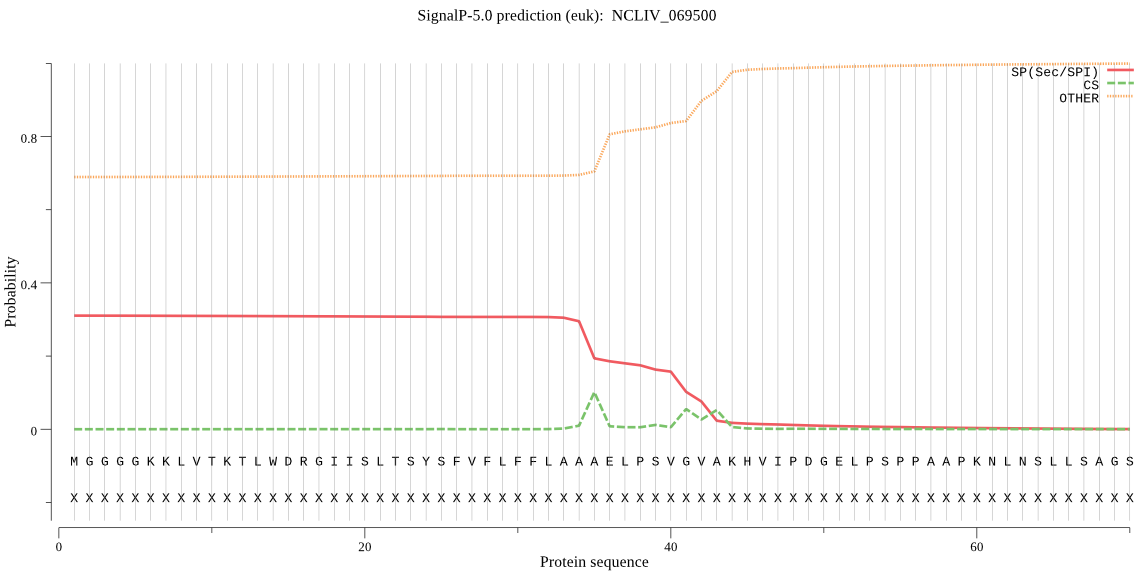
| NCLIV_069500 | SP | 0.433785 | 0.562361 | 0.003854 | CS pos: 35-36. AAA-EL. Pr: 0.4773 |
MGGGGKKLVTKTLWDRGIISLTSYSFVFLFFLAAAELPSVGVAKHVIPDGELPSPPAAPK NLNSLLSAGSPVDFTTLFDAASQASFNAWLVAVRRTLHEWPETAYNEYRTSAVIHKLLKE MNIRVTSGWGTNVVGMSEEEAKMARARREGTGLVAEIGTGKEPCVALRADIDALPIFEQT DVPFRSRVDGKMHACGHDAHATMLLGAAALLKQLEPHIEPAEEGGGGALMMREEGVLTAA PPVEFIFGMHVAPTLPTGELSTRKGVMMAAATQFSITVTGRGGHGAMPHDTIDPSPAVSA IVQGLYAIVARETSFTEDNAGVISVTQIQGGTTFNVIPSEYFIGGTVRTLHMGTMRNLKA RVVDLVESFAKAFRCQARVQYGSVDYVPLVNDPEATETEDPTMGGEDFAFFLENVPGTFA WIGTGSGAEDQPGHVPTNKALHNPEFAVDESVLSRGAALHAFVALRTFSFLANKKHIGAS EPRTTCGAGRKSGQRLEDASCGVQEPSK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NCLIV_069500 | 137 S | VVGMSEEEA | 0.996 | unsp | NCLIV_069500 | 492 S | AGRKSGQRL | 0.992 | unsp |