

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
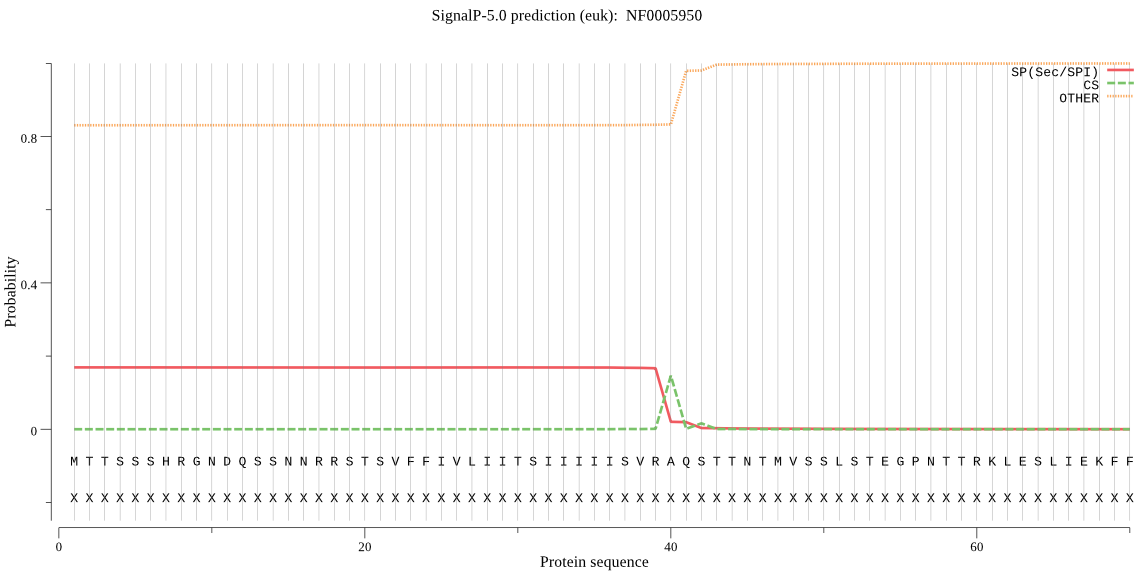
| NF0005950 | SP | 0.431973 | 0.556953 | 0.011074 | CS pos: 40-41. VRA-QS. Pr: 0.9102 |
MTTSSSHRGNDQSSNNRRSTSVFFIVLIITSIIIIISVRAQSTTNTMVSSLSTEGPNTTR KLESLIEKFFSKSDPKASNTAHKHSNNWVVLIDTSRFWFNYRHVANTLSIYHIAKNLGIP DSQILLMLADDVACNPRNRFPGEVFNNKNRQKNLYGENVEVDYRGYEVTVENFFRVLTGR HHEAVPTSKRLMSDEHSNVLLFMTGHGGDEFFKFQDQEEINSADIADVIHQMSERRRFRE MLMIVDTCQAGSLFDKLYTPNVIAVGSSKRGENSYSHHSDLDIGVAVIDRFTYSSLEFFE NRDFRHYSPSLRNWFNSYTAESLHSTPNMRYDLYPRNLDTVPITDFFGSDIKIEFQDEQY PRGNATLLSSYKPVHTNEEYKIAFPSSNDTAVEAQETEVTFENNLSIVNKNIIALFIFGV LICLFLK
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0005950 | T | 2 | 0.536 | 0.239 | NF0005950 | T | 3 | 0.516 | 0.120 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0005950 | T | 2 | 0.536 | 0.239 | NF0005950 | T | 3 | 0.516 | 0.120 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0005950 | 71 S | EKFFSKSDP | 0.996 | unsp | NF0005950 | 71 S | EKFFSKSDP | 0.996 | unsp | NF0005950 | 71 S | EKFFSKSDP | 0.996 | unsp | NF0005950 | 193 S | KRLMSDEHS | 0.993 | unsp | NF0005950 | 6 S | TTSSSHRGN | 0.994 | unsp | NF0005950 | 42 S | VRAQSTTNT | 0.994 | unsp |