

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0023630 | OTHER | 0.999993 | 0.000003 | 0.000004 |
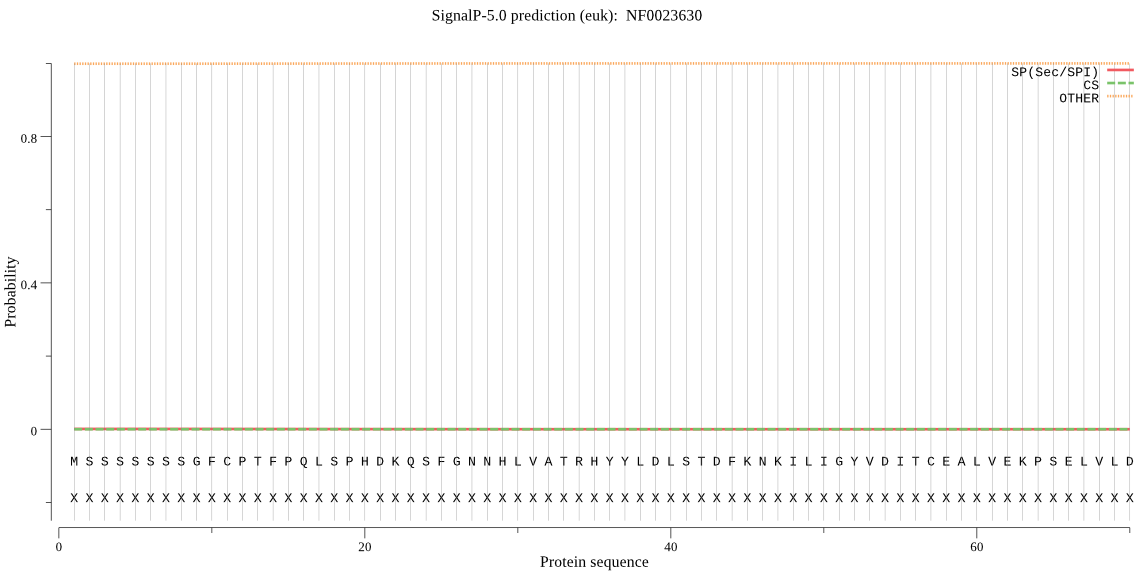
MSSSSSSSGFCPTFPQLSPHDKQSFGNNHLVATRHYYLDLSTDFKNKILIGYVDITCEAL VEKPSELVLDARNLNIVQTKFLYKKHHKHHEESVNEQQQQEMEVSFQQFSKKEVSEKGYP AVFGQPLVINLGASERIDLKKGDLFVVRVFYETTQESEAIQWLNPEQTLGKKHPYLFTQC QAIHARSFLPCQDTPSNKITYSATIRTEKPLVAAMSAIKQWEDDTSHINIYHFQQNVPIP SYLIALVVGALESKQIGPRSTLYCEEEALEKAAEEFSQTEDFIKAAEEFLPPYAWGKYDL VVLPGSYPFGGMENSNLTFLTPTLLAGDKSLANVVAHEIAHSWSGNYTTNATWEEFWLNE GFTVFIERRILARLNGEEYAKFHAQIGYASLTASINHYKEINQTQFTKMIPKLDNIDPDD AFSSVPYEKGFNFLYYLADQIVGDISRFEKFLYHYFTLFAQKTVTSQQMKECFLDFFSDV EKTKEIDWDAWFYSEGDVIVKNNFENSLSVAANELCKRWVEAGEDASSFSKSDIAGWTST QLSYFLDILLENEKPLNIQKLDEIYDLSKYTNSEIRNGWQLLALKHKYSPIKSQVVDFIT SQGRMKFVRPLYRELFKFDKELAVSTFLEHKNFYHAVARKMIEKDLQLDRPNAAVKQ
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0023630 | S | 4 | 0.550 | 0.018 | NF0023630 | S | 3 | 0.528 | 0.029 | NF0023630 | S | 2 | 0.523 | 0.078 | NF0023630 | S | 5 | 0.522 | 0.029 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0023630 | S | 4 | 0.550 | 0.018 | NF0023630 | S | 3 | 0.528 | 0.029 | NF0023630 | S | 2 | 0.523 | 0.078 | NF0023630 | S | 5 | 0.522 | 0.029 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0023630 | 530 S | ASSFSKSDI | 0.995 | unsp | NF0023630 | 530 S | ASSFSKSDI | 0.995 | unsp | NF0023630 | 530 S | ASSFSKSDI | 0.995 | unsp | NF0023630 | 18 S | FPQLSPHDK | 0.994 | unsp | NF0023630 | 115 S | KKEVSEKGY | 0.996 | unsp |