

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0028030 | SP | 0.023375 | 0.976587 | 0.000037 | CS pos: 25-26. TLA-AT. Pr: 0.8274 |
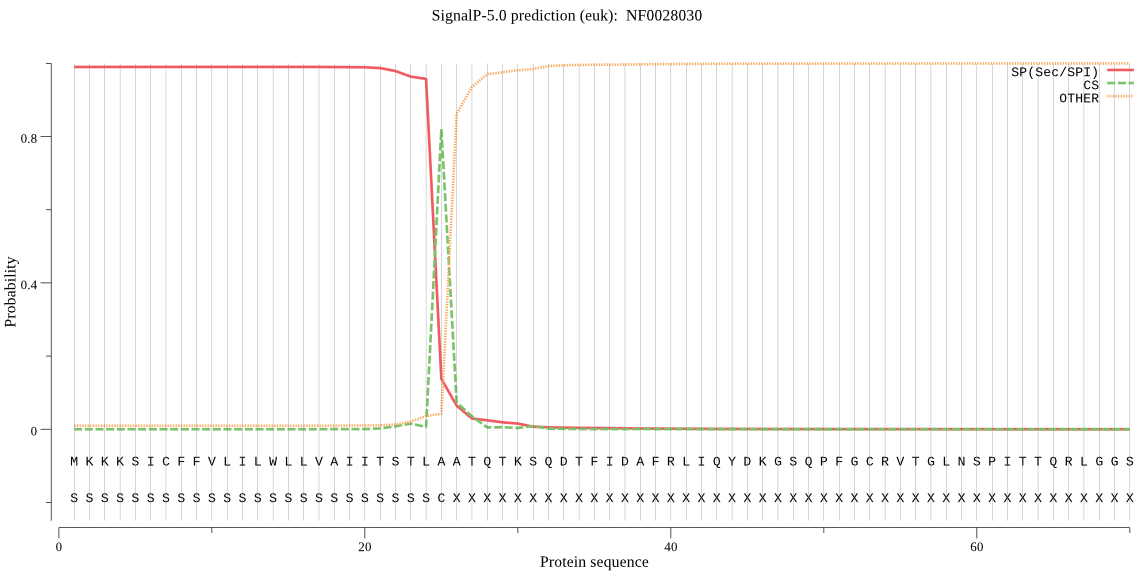
MKKKSICFFVLILWLLVAIITSTLAATQTKSQDTFIDAFRLIQYDKGSQPFGCRVTGLNS PITTQRLGGSSALSAARNVIMMKIEELLSSNKTSVQSILNREDVSGLLIVLPPKDDEISP NSIDQSRNIEKQFLVSTFDKPIYFIFQNEKTSEMLSYFENLSEFSLLSESYQAVVSEGEA KPIDQIPVVNIFSTLQGKQTEKSQTIGIVAHYDSVSSIPGLATTFNADGSGMIGLLEVSR LFQKLYSIPSTKASHNLLFILTGGSRLGFAGTKHWIKQTGPRLVNQLDFVLCLDSLTSRV NSRDDESFKLFMHISEKTNDKNVINLYKHFVSTAKQLQIGLQFIEKKIDRELVWEHEVFA KKGIVSATISAQSVPSSSMLTRTNSLDISKVDIDQLRKSIKLVSEALARHLYDIDHNIRF FEDENDKFIRAFIKNVENTSRFTPTLSKESDLVKTLKLTLQQFTSNDQTVQTQDFLLSNS AYKFYTNIKVKLSLLKVTPITFDLILISSVIIYLLFLHIVLVGPKQVLFNIASLFEKQER KRK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0028030 | 302 S | SRVNSRDDE | 0.998 | unsp | NF0028030 | 399 S | QLRKSIKLV | 0.99 | unsp |