

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0048890 | SP | 0.207802 | 0.791283 | 0.000915 | CS pos: 18-19. LLG-VY. Pr: 0.7732 |
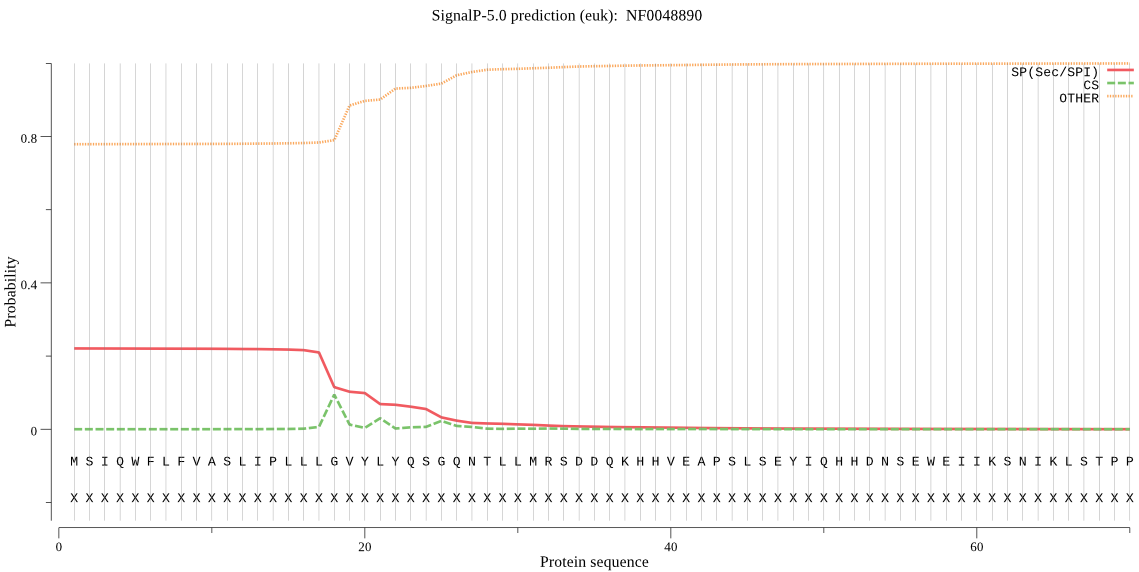
MSIQWFLFVASLIPLLLGVYLYQSGQNTLLMRSDDQKHHVEAPSLSEYIQHHDNSEWEII KSNIKLSTPPSTIHHLRHKRTKLEWFHVDHPNDHNNFFSISFRTPPQDHTGLTHILEHLV LCGSKKYPVRDAFFSMLKRSLSTYMNAWTAADMTSYPFGTQNQKDYYNLMQIYLDAVFNP LLRKLDFTQEGYRFEFQEHDNLQSNLTFKGVVYSEMQGVVGDANNYFSFELNRLLFDRFK NFTSLQHSDTISKFLKSLHSYSFNYGGKPENIIDLKYEDLDAYHKKYYSPNNALIFTYGS FSLIDHLNFVKNQQILDQFEEVMDVTSAPQDEVNKLDDQKHHSSILQQHLQSMITQSNEI IYFYGPEESIVYNPEKQLKYAMTFFTNQPANGFFEVFQMRMLDLLLLENPRGPLYQSLIE SKLAPRFLSITGFEKTDLLRKQPTFVIGVSEIGESDVSIIEETINKTLSSVIKEGFRKEY VESILNRIEISNKRVSANYGTSLMMEIPPFWCNGDSHRLEEVIDSEMNIKRLRQELEKSP TLFQDLISKYLRTDLALRVVMRPNKNQLNEFKEFEQAVLQTTQNNMSESEKKQIISHSIQ LQEYQNAEKLKDVDAILPSLTLSDIDTSILYQLDPHIQNFQIMGINVTTYEQETNGLIHL SVNVELWDGEKILLDKQLIDLLPFFTYCLTKMGTRELDKKELTEKIDMFTGGITSNLFFS PKAIPSPRTCIEKFGVNLYSEAVYEKMPIMFKLLLDIMTSPRFEDVKYLESLFEEYATSI SESLTQNGHEFSRSYSASVLNDYSLLTERLTGVSHFIFIKEFKKIHQANLAEVLQERMKL LLHSIYHHMTIAITCEHGHLQDLD
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0048890 | 587 S | QNNMSESEK | 0.995 | unsp | NF0048890 | 587 S | QNNMSESEK | 0.995 | unsp | NF0048890 | 587 S | QNNMSESEK | 0.995 | unsp | NF0048890 | 623 S | SLTLSDIDT | 0.993 | unsp | NF0048890 | 302 S | YGSFSLIDH | 0.996 | unsp | NF0048890 | 458 S | ESDVSIIEE | 0.993 | unsp |