

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
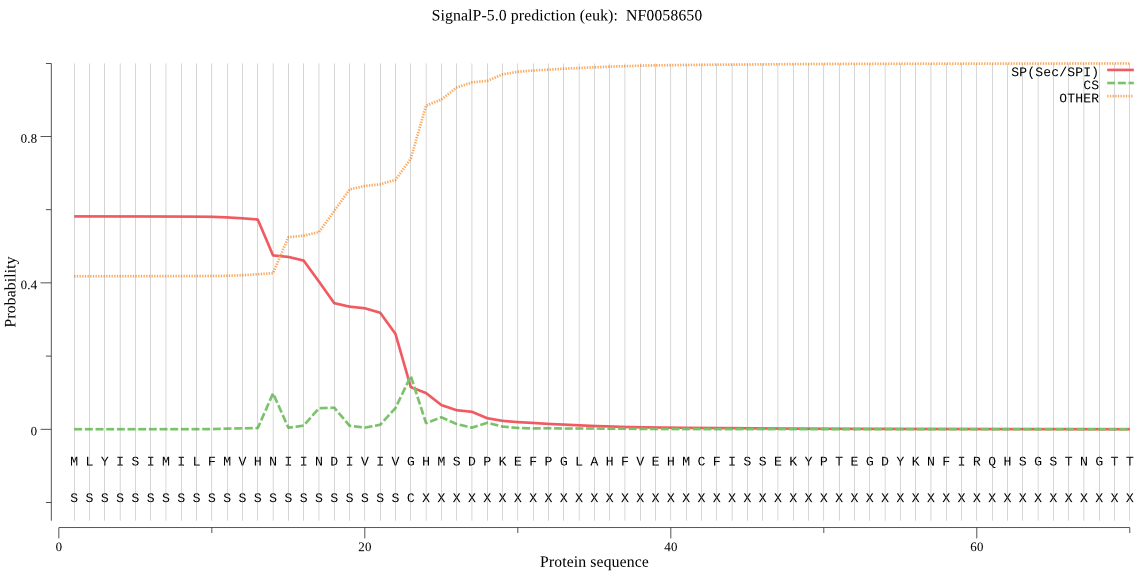
| NF0058650 | SP | 0.123020 | 0.876918 | 0.000062 | CS pos: 17-18. IIN-DI. Pr: 0.3456 |
MLYISIMILFMVHNIINDIVIVGHMSDPKEFPGLAHFVEHMCFISSEKYPTEGDYKNFIR QHSGSTNGTTTAEHTNYYFTISNEYLEEALDRFAQFFISPLFAENAIVREVEAIDKEFKK NFQIDSRRLYQLMKESCNENHPFKKFGTGNVQTLKIDPEANHLNVREHLVNFFNRYYSSN QMKLCVIGNYSFQEMEKLVRNSFSAIKNSHISKHEFYPPSDATKPFQPSDLAKLFKYEPV TDTTDLTLLFPLSIDCPKIIQRRNMYYKSSSIYCLLQLFGHEGKGSLYSLLKHKSWAESL TSYTYDYNGVNDPGKQFSLFAIKIKLTKTGINFWKEIVEHVFEYIQILKTQGVPRWIFEE FSHLQQLAFDNAQFTSSYASTLSESLQQYLPEDVIGANYLLYEFDEDHTNYILSHFHPDN LNIYLSSKTFSDLDSSERWYGFKYRKGDLETNFISTLRNKKNETPQLHLPFKNTYIPYNL GLVTDTNPITQYPVKIVDRPKIKTFFKKDDYFNTPRADIIVLLTIPQSFEAPLNVVYVEL FVDTVRYLLNEELYMASLAKIASNITNVERGVEVHVNGLREHLVDVIMVILEEIVKAADD RNHLTQHVFDYVKQNNLRNHQNKKFSFQSHEIATYESSRLFLQRKFTFFEFAQALERVEI DEFKNFVKSWLTVLRIECLIHGNFTKDNALHISQQVEKIMFDNRMNANKILTPLPSQEYL RNVAVNPTNVELVVPILNYDTSNQNNAILVSYQIGYRSLRSDCLLELFTQICSSKYFTHM RTQKQYGYIVMCYHRFIHNVSFLSCLIQTTTHELTDILVENDHFFKNNVSKLLEELTEEQ FNDIVGSIIQKALEKEKTVQQRSQRFWNEIENRQLKFNRYELKAEQFKKFTKKDINEFYE NFVNGPHCKKSVFLVSNKTYHKDQFSNPNIVLVEDIAQFKSTLEYHCSISEF
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0058650 | 431 S | SKTFSDLDS | 0.993 | unsp | NF0058650 | 511 Y | KKDDYFNTP | 0.99 | unsp |