

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0062180 | SP | 0.419383 | 0.578334 | 0.002283 | CS pos: 42-43. VHS-EI. Pr: 0.4298 |
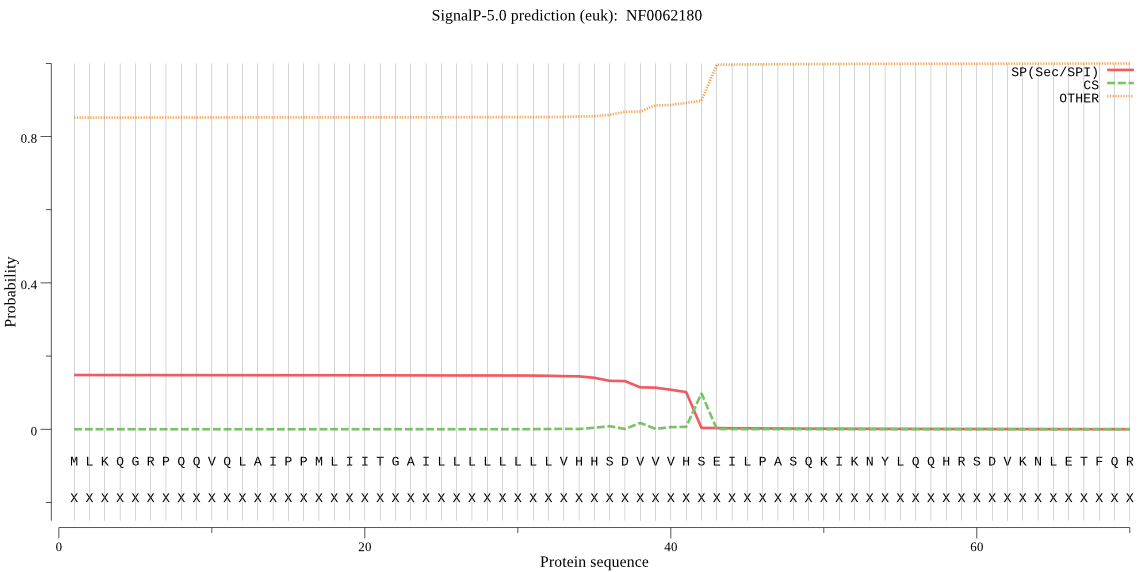
MLKQGRPQQVQLAIPPMLIITGAILLLLLLLLVHHSDVVVHSEILPASQKIKNYLQQHRS DVKNLETFQRWVRYASLPEHEKDIQLDIAKYLKEELHFNKVDIWTMDQQKVELENSKFFN TPRNFDSLKKSPIVVGVLDGTAPEDLVETTQEGSIQNVASTKFKPYKSLIINGHIDVVPV GDPKQWYLENPFSGHINDSKIYGRGTTDMKGGLYSGLLAIEAIQKSLGVTNMKGKIIVHS VVEEESGGAGTVSTILRGYGHADAGIFPEPSNFLIFPQQQGSLWFRITVYGKSAHGGTRY DGISAIEKSQIVLNAIKKLEDERTDLIRNVLKNKLFENITIPVPINVGVIRGGEWPSSVP DFTVIEGRFGIIPNYETVEDAKQVLHDLVFKTIPELDKAHFSAYPCKLEFIGASWVPGYV PLEHEFVSQLSRSFSEVMGQNPIIASSPWATDAGYVNMMGNTPSVVFGPGVTHMAHQTNE YIPIDNIYKAAEIIALTIVEWCGV