

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0067420 | OTHER | 0.999153 | 0.000801 | 0.000046 |
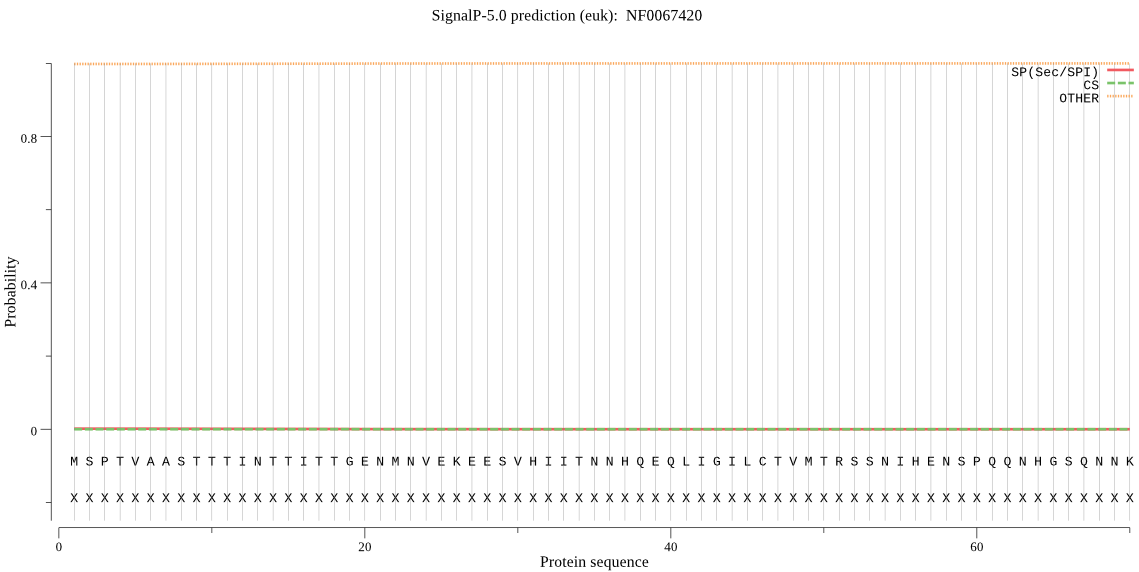
MSPTVAASTTTINTTITTGENMNVEKEESVHIITNNHQEQLIGILCTVMTRSSNIHENSP QQNHGSQNNKKPIAVLCHGLACHKNYFIFPRMESNEYDTFRFDFSGNGESDGEFSYANYH KEVEDLHCVINYLKEKLNYSKISIIGHSKGGNVVLQYASKYPQELQHGVVVNISGRFDMS KTPMNRFSESERHQLENEGIFLWKVFTLEPSLCVRKKQEITTPTNNEEGLTRRYYVTKEA LQERESLNTKIEFNLLYPLKLYSIHGDNDDVIPFQDSQEFHEFLLEQMRKRNLIDRFSHQ VICVKGANHFFAGHYETLIGILKDLLAKE
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0067420 | T | 4 | 0.682 | 0.381 | NF0067420 | S | 2 | 0.621 | 0.076 | NF0067420 | T | 9 | 0.594 | 0.105 | NF0067420 | T | 10 | 0.574 | 0.040 | NF0067420 | T | 15 | 0.546 | 0.104 | NF0067420 | T | 17 | 0.543 | 0.081 | NF0067420 | T | 11 | 0.539 | 0.058 | NF0067420 | T | 14 | 0.534 | 0.041 | NF0067420 | T | 18 | 0.522 | 0.082 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0067420 | T | 4 | 0.682 | 0.381 | NF0067420 | S | 2 | 0.621 | 0.076 | NF0067420 | T | 9 | 0.594 | 0.105 | NF0067420 | T | 10 | 0.574 | 0.040 | NF0067420 | T | 15 | 0.546 | 0.104 | NF0067420 | T | 17 | 0.543 | 0.081 | NF0067420 | T | 11 | 0.539 | 0.058 | NF0067420 | T | 14 | 0.534 | 0.041 | NF0067420 | T | 18 | 0.522 | 0.082 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0067420 | 188 S | MNRFSESER | 0.991 | unsp | NF0067420 | 277 S | PFQDSQEFH | 0.992 | unsp |