

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0068180 | SP | 0.000087 | 0.999909 | 0.000004 | CS pos: 20-21. VLA-QE. Pr: 0.8671 |
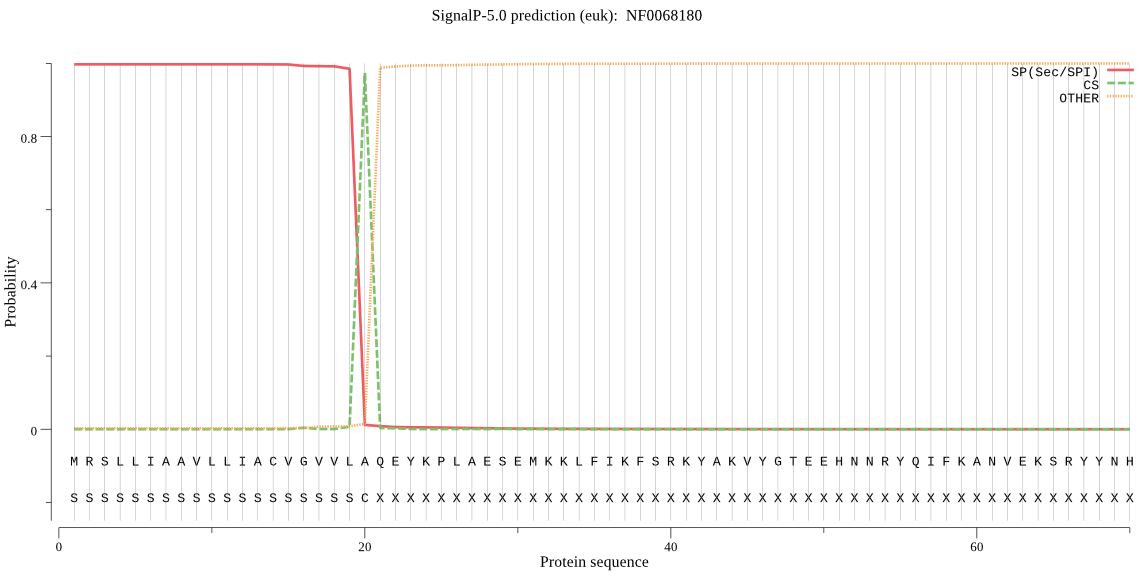
MRSLLIAAVLLIACVGVVLAQEYKPLAESEMKKLFIKFSRKYAKVYGTEEHNNRYQIFKA NVEKSRYYNHVGKRENFGITKFSDLTPEEFKRMFLMKTYTPEEAKKILAAPQHAVLSEKE VQTAPTSFDWRQHGAVTRVKNQGACGSCWTFSTTGNVEGQWAIKKGKLVSLSEQQLVDCD HNCVTYQNQQACDSGCNGGLMWSAFQYVIKNGGLDTEDSYPYEGVDDTCRFNKSNVAATI SSWTSISSDENQMAAWLAANGPISIAINAEWLQYYTSGISDPWFCNPQDLDHGVLIVGYG VGKSWLGSEENYWIVKNSWGSEKNEDGYFRIIRGKGKCGLNSVPSSSIV
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0068180 | 321 S | NSWGSEKNE | 0.995 | unsp |