

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0069190 | SP | 0.388005 | 0.603236 | 0.008759 | CS pos: 26-27. VSC-RV. Pr: 0.5998 |
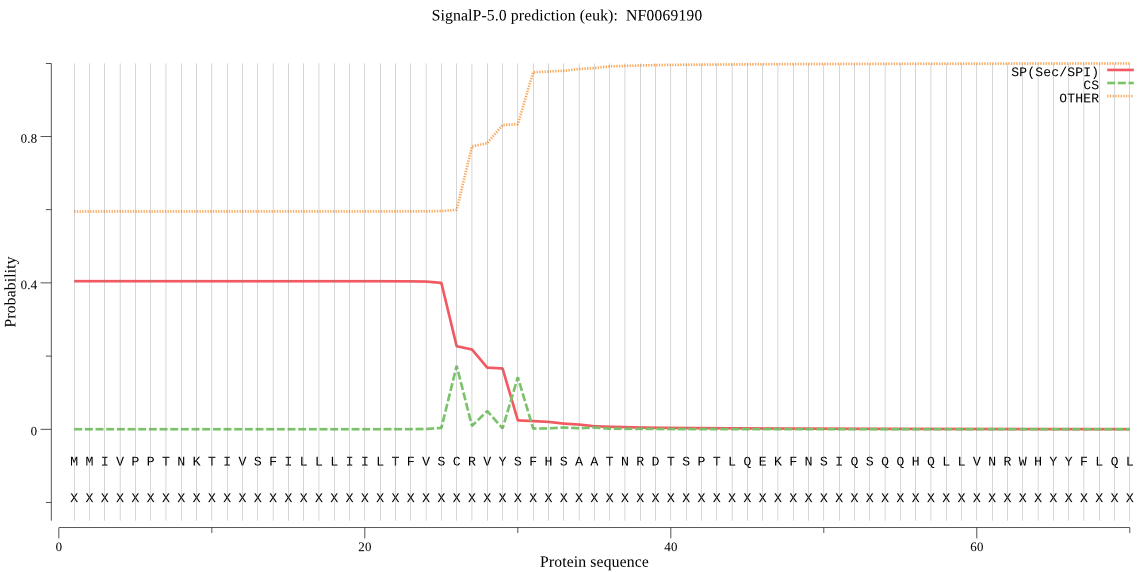
MMIVPPTNKTIVSFILLLIILTFVSCRVYSFHSAATNRDTSPTLQEKFNSIQSQQHQLLV NRWHYYFLQLINQVNSYKYNCANIRNDPNDDEKNHSENQSNRESILELKQTWFAAPNELT QFPQHYLKLIQNGLLSSREDLWKRFSSLMTTIKMSKIGEFGREHLRVLNELLKRAEQRDL LRMVKLRNVNNLEELRKRQRAMNIPETFDSRMQWPSCIHPIRDQGNCGSCYAFASTEVMS DRFCIFSNASVNVVLSPQDLISCSWYSFGCDGGIPGLVFDYLHKDGVVSDACLPYLSYNG NTGHKCPSYCFNNASKPLKDDKYFAKSVYHVGSFIENKEDRVIAIQQEILKNGPVNADFM VFQDFVTYKSGVYRHLTGSFAGIHAVKIIGWGVENNIPYWLVANSWGTNWGFDFGHFKIL RGQDECGIEFSVYSGIPNLENVPKTTTPK
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0069190 | T | 446 | 0.606 | 0.249 | NF0069190 | T | 447 | 0.603 | 0.160 | NF0069190 | T | 445 | 0.575 | 0.111 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0069190 | T | 446 | 0.606 | 0.249 | NF0069190 | T | 447 | 0.603 | 0.160 | NF0069190 | T | 445 | 0.575 | 0.111 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0069190 | 41 S | NRDTSPTLQ | 0.998 | unsp |