

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
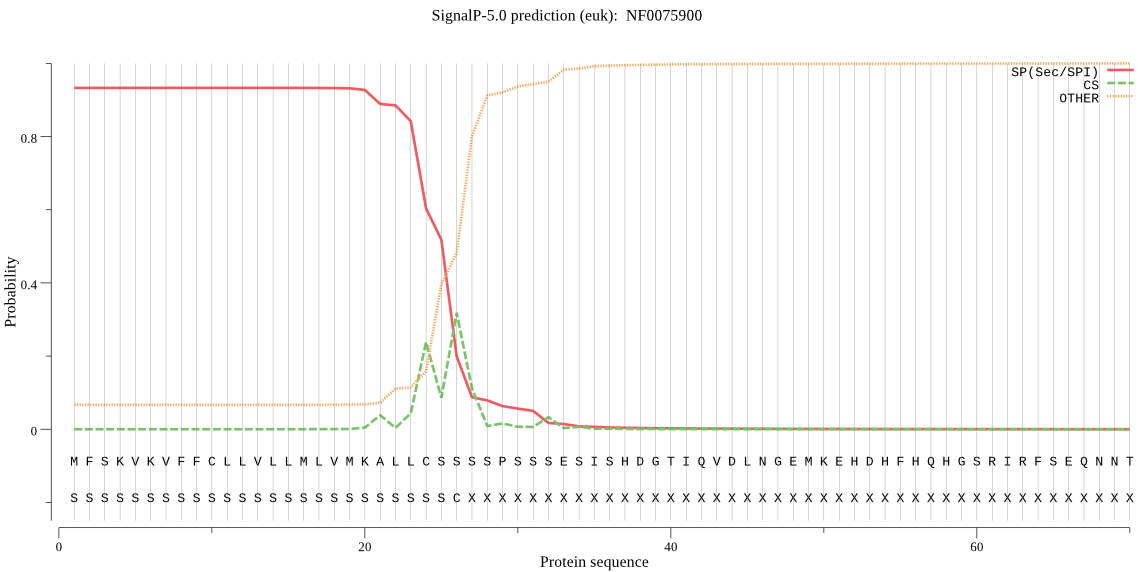
| NF0075900 | SP | 0.024250 | 0.974656 | 0.001094 | CS pos: 24-25. LLC-SS. Pr: 0.3626 |
MFSKVKVFFCLLVLLMLVMKALLCSSSSPSSSESISHDGTIQVDLNGEMKEHDHFHQHGS RIRFSEQNNTWMRKRLEERFLKYVQIDTQSDPHSTTVPSTEKQKVLAHLLLEELKTMKGL KNVDMDQYGNVYATLPKNLERDVLSVGFLAHMDTATEISGFNVTPCVHRNYKDGENLIVN NLTIVDYKIDGKELLKMIGHDIVTASGGTLLGADDKAGIAEIMTAIEFLIENPSIEHGDI QIGFTIDEEIGRGPHYFDIPKFSAAVAFTVDGGEKGTYNVESFSADLVTVAFTGFNIHPG TAYSRMINSMKVAAQFLDRVNEQMPSPDISKDREGFAHCLDINGNVEKTTIRCILRSFEE SELKQYSQEISKIIDQLRFEHSNVTIEMNVVQQYRNMKQVLDKYPFMELSIMEVYSALNM KDIVSIPIRGGTDGSQLSWMGLPTGKFFHPYSNSTTRNSLILLHHNNTANIFAGGHNFHS QSEFVSLHDMADSVKVIVMLSQTLSKVPPLPIMKGHH
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0075900 | 99 S | TTVPSTEKQ | 0.992 | unsp | NF0075900 | 99 S | TTVPSTEKQ | 0.992 | unsp | NF0075900 | 99 S | TTVPSTEKQ | 0.992 | unsp | NF0075900 | 357 S | CILRSFEES | 0.994 | unsp | NF0075900 | 30 S | SSSPSSSES | 0.996 | unsp | NF0075900 | 31 S | SSPSSSESI | 0.995 | unsp |