

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
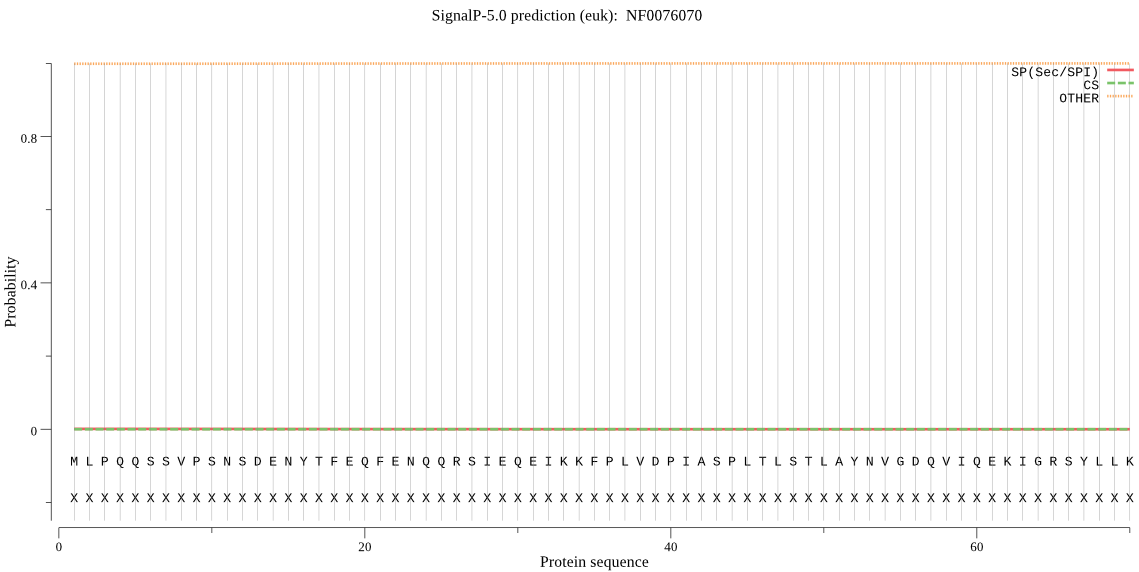
| NF0076070 | OTHER | 0.999942 | 0.000032 | 0.000026 |
MLPQQSSVPSNSDENYTFEQFENQQRSIEQEIKKFPLVDPIASPLTLSTLAYNVGDQVIQ EKIGRSYLLKKYSHIRGIRGDGNCFIRAFVFGLLDFLIRKKDKTLTQQMIDIIDQHYKYL IEVLNYPEHCLEDFYSITKEVFEKVRDGVIHTVEGLLEHYFSDKTTDLMMTSQAIVCFMR FLLSGHIQMNCDDYLPFFLDQYATAKEFCHAEIEIMDRDCDNIHLMAMTSAFKVCIRVEV LDAQSRASDKFDVFGYTIPECSDKEPVVYLLFRPGHYEIIYPRENQ
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0076070 | 248 S | QSRASDKFD | 0.997 | unsp | NF0076070 | 262 S | IPECSDKEP | 0.994 | unsp |