

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
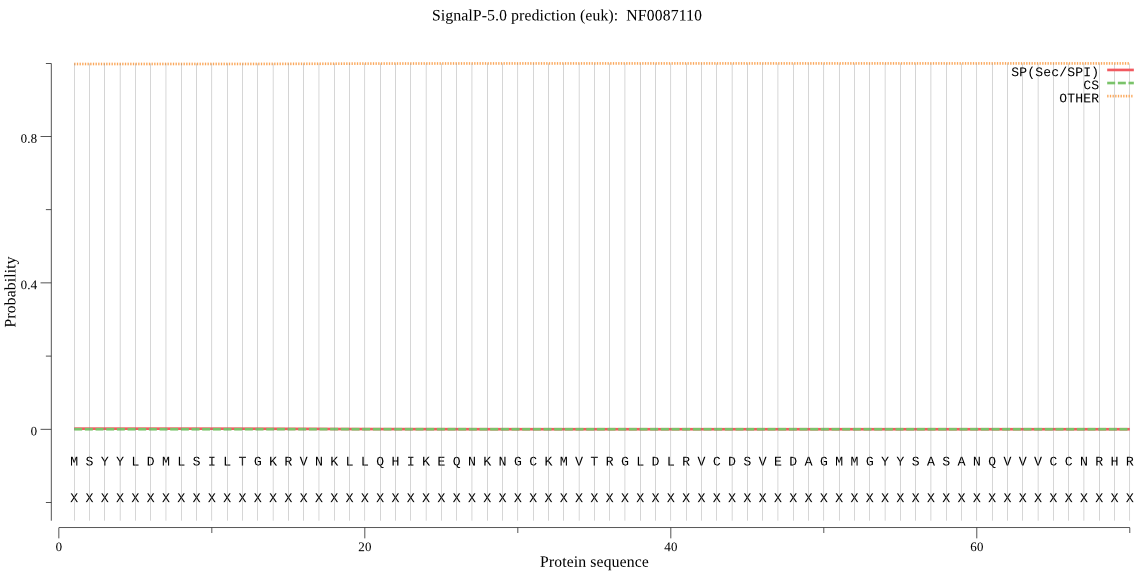
| NF0087110 | OTHER | 0.997322 | 0.002427 | 0.000252 |
MSYYLDMLSILTGKRVNKLLQHIKEQNKNGCKMVTRGLDLRVCDSVEDAGMMGYYSASAN QVVVCCNRHREPEELENTVVHELIHAYDFCRFRQLFYCKVRACSEVRAYSLSGSCSSPDE LSGFTSKEQCLKARAIASTIMACKKSAVSMFMTFSIRVSMMSILSRVRLIIMC
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0087110 | 45 S | RVCDSVEDA | 0.994 | unsp |