

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
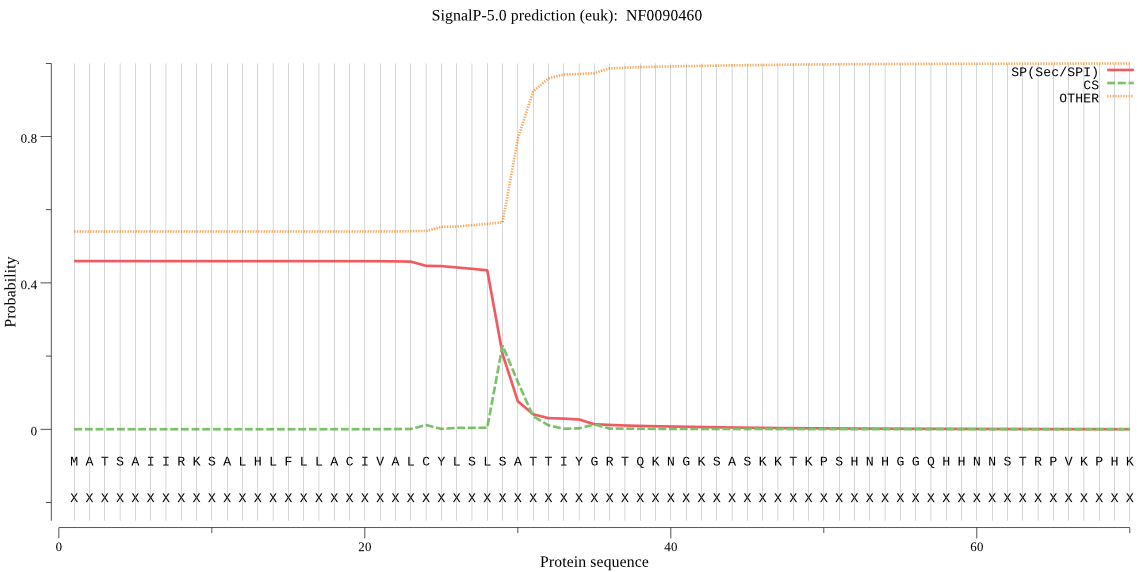
| NF0090460 | SP | 0.317456 | 0.655693 | 0.026851 | CS pos: 29-30. SLS-AT. Pr: 0.6184 |
MATSAIIRKSALHLFLLACIVALCYLSLSATTIYGRTQKNGKSASKKTKPSHNHGGQHHN NSTRPVKPHKPEVVNGTTPANDDDLIERINTNHTIGWKATKYARFENMTIGHLRNSLFGL SLLPNDPDTPRIEGEVRVQLPASFDARTAWPRCVLPIRDQQSCGACWAFSSAYVLAQRYC VATGANSYTVLSPEYQVDCDSLERGCQGGYLSSTWRFLTNTGTVPDTCIPYVSGRTTSAG SAQSCQSQCKDGSALRFYKAKSASIIRGVDQIKIAIMTYGSVQAGFTVYRDFLNYKSGVY KHVSTTALGGHAVALIGWGSENGTPYWIAVNSWSTNWGERGYFRIALGSNESGIESQVYA GLANV
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0090460 | T | 227 | 0.511 | 0.020 | NF0090460 | T | 63 | 0.502 | 0.042 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0090460 | T | 227 | 0.511 | 0.020 | NF0090460 | T | 63 | 0.502 | 0.042 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0090460 | 192 S | YTVLSPEYQ | 0.992 | unsp |