

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
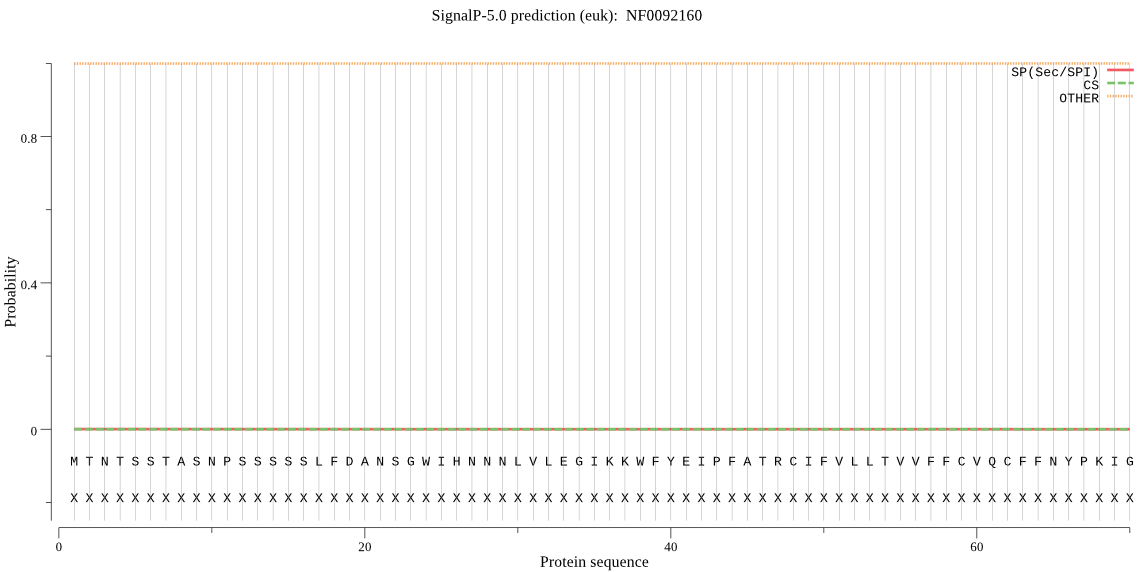
| NF0092160 | OTHER | 0.999999 | 0.000000 | 0.000001 |
MTNTSSTASNPSSSSSLFDANSGWIHNNNLVLEGIKKWFYEIPFATRCIFVLLTVVFFCV QCFFNYPKIGDVCLSYLDITENHYSDMYRIVSYALFHANLMHYLFNIMAQISLSSLLETH RFKSSIKLFACTFISIVFASISQLVIAWLMDTISLYVFGYNVRSGNFVKGSNKSASSMFN MLNDIPIINECSIGFSGVIFTYLTILSIDGFQSTQSLFGLVTVPSKYYPWILLFVTELLI SNASFVGHLCGIVMGYFYVFVFERNVLISHYYEKLLCQTENMIIPNVVINLECYWPNSQQ KTNLAPSNNSGSFLAHMMETGQVSSSYARSGNGDYWEQLSSSGRTLGYH
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0092160 | T | 2 | 0.723 | 0.063 | NF0092160 | T | 4 | 0.686 | 0.027 | NF0092160 | T | 7 | 0.645 | 0.172 | NF0092160 | S | 5 | 0.590 | 0.032 | NF0092160 | S | 6 | 0.547 | 0.020 | NF0092160 | S | 9 | 0.505 | 0.043 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0092160 | T | 2 | 0.723 | 0.063 | NF0092160 | T | 4 | 0.686 | 0.027 | NF0092160 | T | 7 | 0.645 | 0.172 | NF0092160 | S | 5 | 0.590 | 0.032 | NF0092160 | S | 6 | 0.547 | 0.020 | NF0092160 | S | 9 | 0.505 | 0.043 |