

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0099990 | SP | 0.102441 | 0.897391 | 0.000168 | CS pos: 33-34. LQA-LE. Pr: 0.1942 |
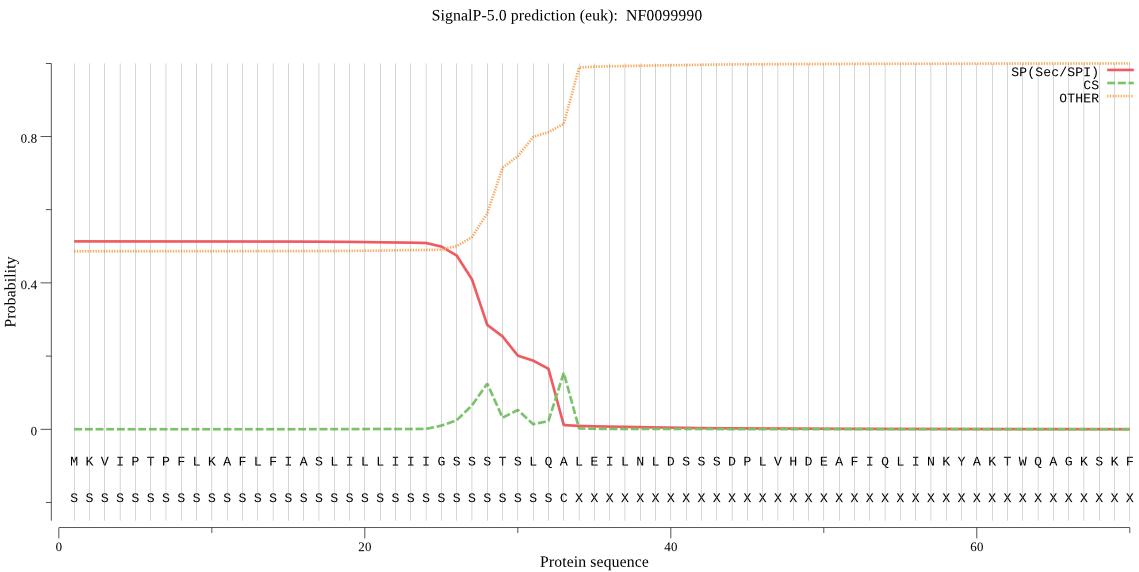
MKVIPTPFLKAFLFIASLILLIIIGSSSTSLQALEILNLDSSSDPLVHDEAFIQLINKYA KTWQAGKSKFFEGKRLSHARRLIGLGLPTPEQRASYPKKNSLMMGEEANSLEKYLVKMDA LPDSYNAANDSNYYMCQQLHRIRNQEQCGSCWAFSISEMVADRFCIGTRGKINTIMSPQW MVSCDTADNGCNGGEFPTAFQFVETTGLVSDGCVPYQSGNGFVPPCPNSCANGEDINVRY RTKNSRNFDVNDMKSVQASILANGPVISGFKVYRDFYNYRSGVYKHVAGGLVGGHAIKVV GWGVTQSNVPYWIVANSWSDEWGMNGYFWILRGTNECSIEENMWETIPAL