

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0105870 | OTHER | 0.999735 | 0.000263 | 0.000002 |
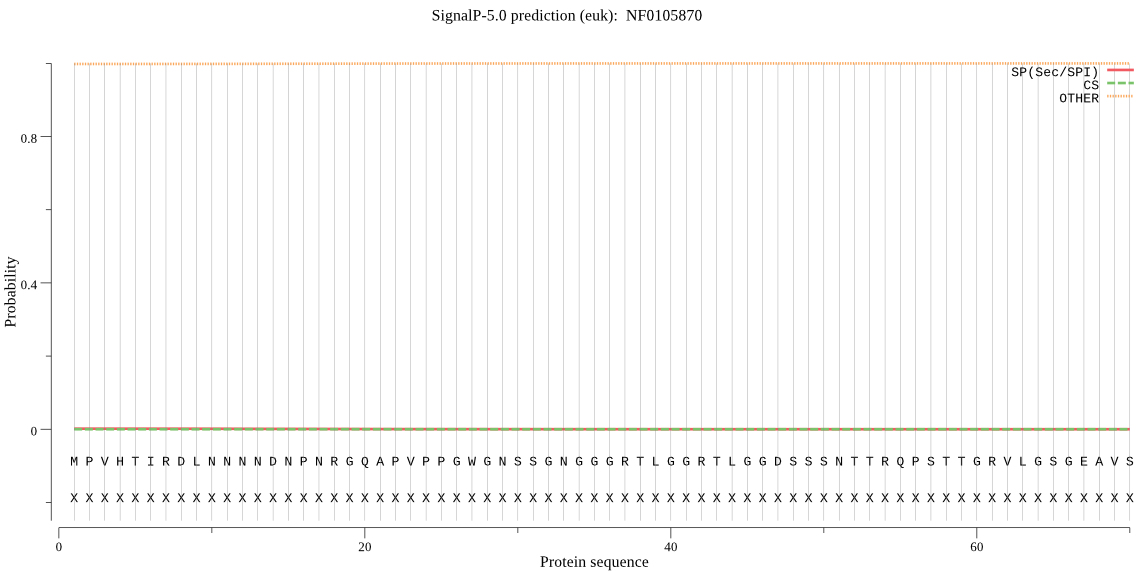
MPVHTIRDLNNNNDNPNRGQAPVPPGWGNSSGNGGGRTLGGRTLGGDSSSNTTRQPSTTG RVLGSGEAVSYQPSSANYADGGAIGGGGGFFSGFRNMFSGFGRSSNTTPSGGSYTATGQP VDVETGGAATISNDPYQNQYNMNEDMFWSYNLHNIQPQWYHMCLMCCCPCFVGPPCSPVR KQDYKNMLMTFCFWISIVQLIYFIVELSVGGFDPQNPSIGPSSLTLLKLGAKSSYYIKEK YELWRLVVPLIMHAGILHIFMNLFIQIMVCMGYEKNWRWYRVIPIYFISGIAGNLLSCVA MPASISVGASGAIMGLIGAKVSNIIVRWTRIPTQQKVMQCINVGIIIFITLLWSFSDYID WAGHIGGLVTGFILGFACFAVTEIQDKVYKWTIFSVSVGLTLVTLLTLSLVFGLVTDTSK YAL
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0105870 | T | 117 | 0.602 | 0.128 | NF0105870 | T | 115 | 0.582 | 0.048 | NF0105870 | T | 58 | 0.537 | 0.088 | NF0105870 | T | 59 | 0.533 | 0.263 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0105870 | T | 117 | 0.602 | 0.128 | NF0105870 | T | 115 | 0.582 | 0.048 | NF0105870 | T | 58 | 0.537 | 0.088 | NF0105870 | T | 59 | 0.533 | 0.263 |
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0105870 | 57 S | TRQPSTTGR | 0.994 | unsp |