

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0108310 | OTHER | 0.999934 | 0.000013 | 0.000053 |
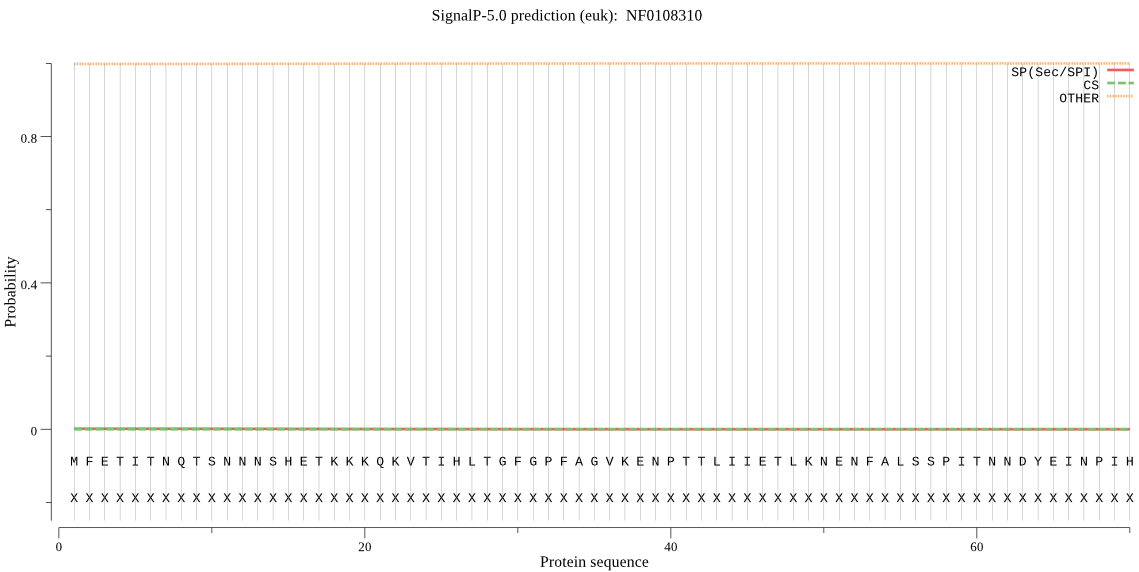
MFETITNQTSNNNSHETKKKQKVTIHLTGFGPFAGVKENPTTLIIETLKNENFALSSPIT NNDYEINPIHYSIIETSMKAVQQYFEEMKCNSGGGDDDSKYPSIDIFIHFGVSGNSIMYE LEERAKNEKTFRARDEQGEQPLNEPINEDLPIDSFLHCNLTTHLNSIIESVNLKLHENSL HPPLSQLVLESARDKLRRINMTGTLGGVKSTKNALSSNPNEDFIQKQLEELRTDLQESLS LITNAGAFCKASQDAGLFICNYCYYHSLQYSTCGDVSRNGLDYNNKKDEMQKKSQRKCYS LFVHVPPHNVIPVKVQVEFVKTLLQVIVESL
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0108310 | 166 S | THLNSIIES | 0.99 | unsp |