

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0109410 | OTHER | 0.993094 | 0.005771 | 0.001134 |
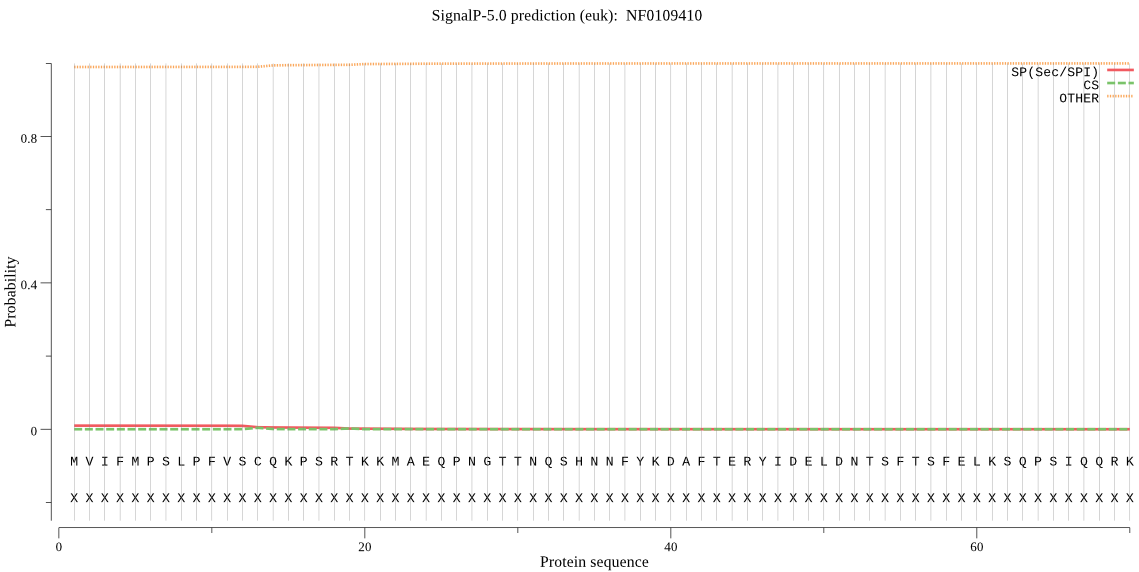
MVIFMPSLPFVSCQKPSRTKKMAEQPNGTTNQSHNNFYKDAFTERYIDELDNTSFTSFEL KSQPSIQQRKPVQNYSVEYDTTKDPNLEDPMDLGEGKSKYTADQTQIDGENQKKKKNRMT DRIISFPYWTALTCVAEVTVFVAMCIVGKGIDDPLNNPMIGPKQEIVIQFGAKDNSLIRA NGSEFWRFFVFMFIHQSVLILLFNLMWLLTTVRKTEGVWKFPRMAVIYMLSGIGGGLLSS VFSFDQISTGSTSCIVGIISASLAELILNWDVVFNPFKLLLSVIMQLLVFFIIGLLPTVD QFAHIGGFICGFLSGVMLCARKQKPDLEKRWAKFTIIVARAFAAVLLVIYFALFFPIFYG LIPMKCEACYWLDPTWAMYFGDNDE