

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0116430 | SP | 0.000984 | 0.999008 | 0.000008 | CS pos: 20-21. VHS-TT. Pr: 0.7388 |
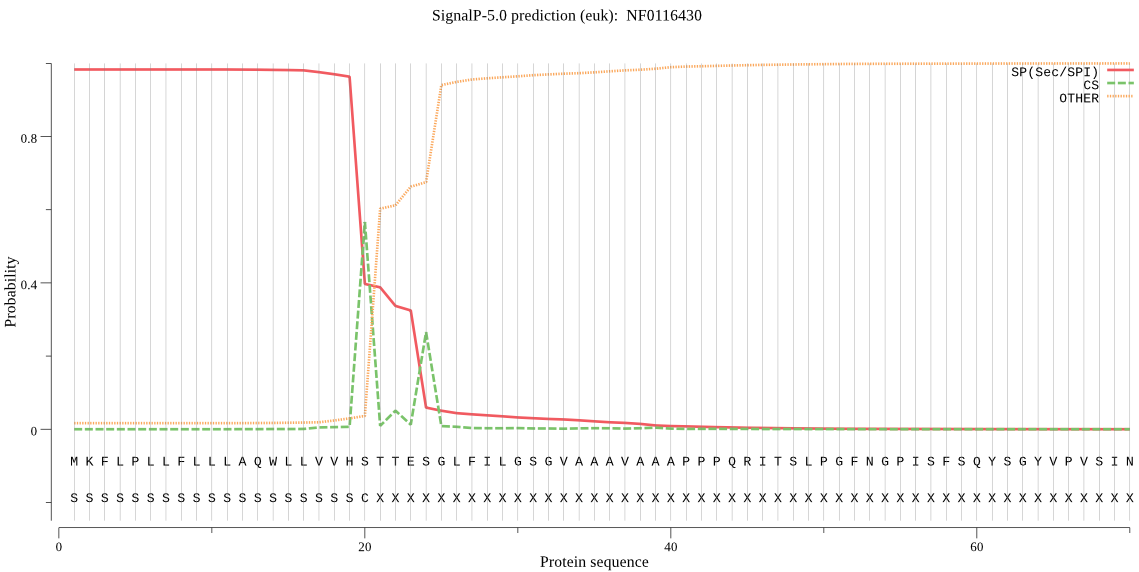
MKFLPLLFLLLAQWLLVVHSTTESGLFILGSGVAAAVAAAPPPQRITSLPGFNGPISFSQ YSGYVPVSINSTYETRFLFYWFVESQSARPSEDPVILWLNGGPGCSSLDGMVTEHGPFLL NNDGKTLRANDYSWNKRANIIYLESPFEVGYSFTTNNNDHVWNDVKSANDVLQFLAIFFS EMFPQYVKNPFYIAAESYGGHYGPTTAVAVLHANQRNQFPFKFNLKGFIVANGIMDDRED TNSIPLFMYQHSLISKSSYDDGLSKCKGDFYKNQNLPECANVISQYYTSIVGVNPYDIYA NCVGDVGPFVDEKSDEFSQKEESFLETLKRNGWMKALTQRPSFKNSNVIIHPLFALSTTR VGSGAPCLAYKPQENWFNLNEVKQALHASPLIPQDHRWQMCNDVVNGNYNRTYDSMIPFY QELLSNGIRGLFYSGDCDLAVNSLGSQNGIYALMKVMNGQIVKPYQSWMMDKQVAGFYQI WSAGSTTMTFKTIKGAGHMVPMTRPKQAQTVLYDFIFNN
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| NF0116430 | T | 47 | 0.501 | 0.322 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| NF0116430 | T | 47 | 0.501 | 0.322 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0116430 | 258 S | ISKSSYDDG | 0.993 | unsp | NF0116430 | 258 S | ISKSSYDDG | 0.993 | unsp | NF0116430 | 258 S | ISKSSYDDG | 0.993 | unsp | NF0116430 | 318 S | SDEFSQKEE | 0.997 | unsp | NF0116430 | 71 S | VSINSTYET | 0.993 | unsp | NF0116430 | 257 S | LISKSSYDD | 0.992 | unsp |