

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
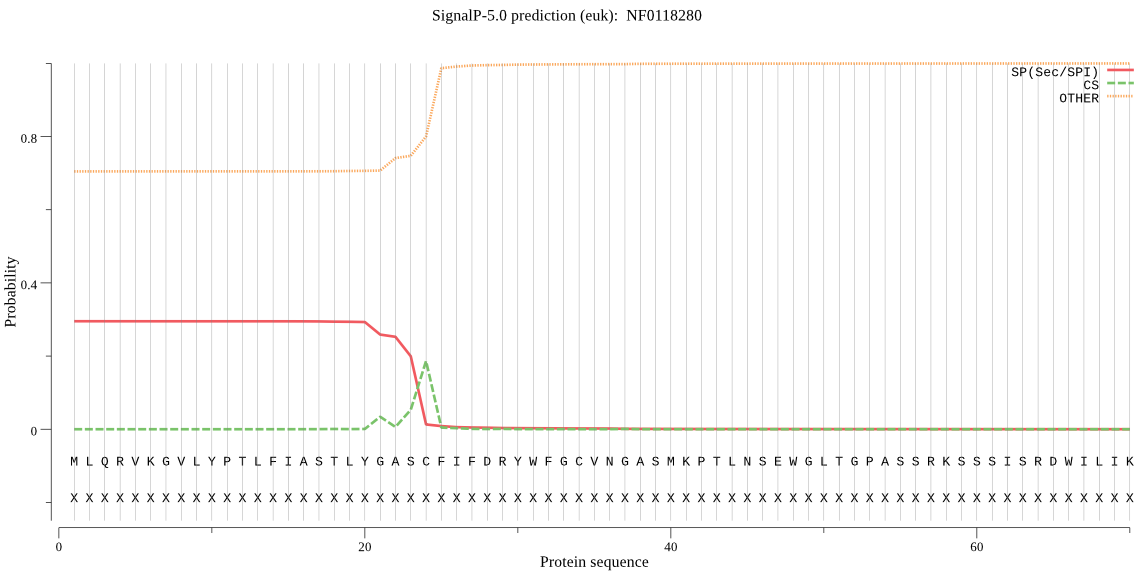
| NF0118280 | SP | 0.362274 | 0.564253 | 0.073472 | CS pos: 24-25. ASC-FI. Pr: 0.7100 |
MLQRVKGVLYPTLFIASTLYGASCFIFDRYWFGCVNGASMKPTLNSEWGLTGPASSRKSS SISRDWILIKRVKDHKNELDINDIVVFVSPEEPRTTAIKRVKALPGQVYQPSRRDTDGHA ASGKYIPKGHVWVEGDNAVSSVDSNMYGPIPMGLIRGKAVCVIYPKFKWL
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| NF0118280 | T | 51 | 0.528 | 0.071 |
| ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|
| NF0118280 | T | 51 | 0.528 | 0.071 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0118280 | 60 S | SRKSSSISR | 0.993 | unsp | NF0118280 | 60 S | SRKSSSISR | 0.993 | unsp | NF0118280 | 60 S | SRKSSSISR | 0.993 | unsp | NF0118280 | 89 S | VVFVSPEEP | 0.998 | unsp | NF0118280 | 112 S | VYQPSRRDT | 0.994 | unsp | NF0118280 | 56 S | GPASSRKSS | 0.995 | unsp | NF0118280 | 59 S | SSRKSSSIS | 0.996 | unsp |