

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0121710 | OTHER | 0.999991 | 0.000002 | 0.000006 |
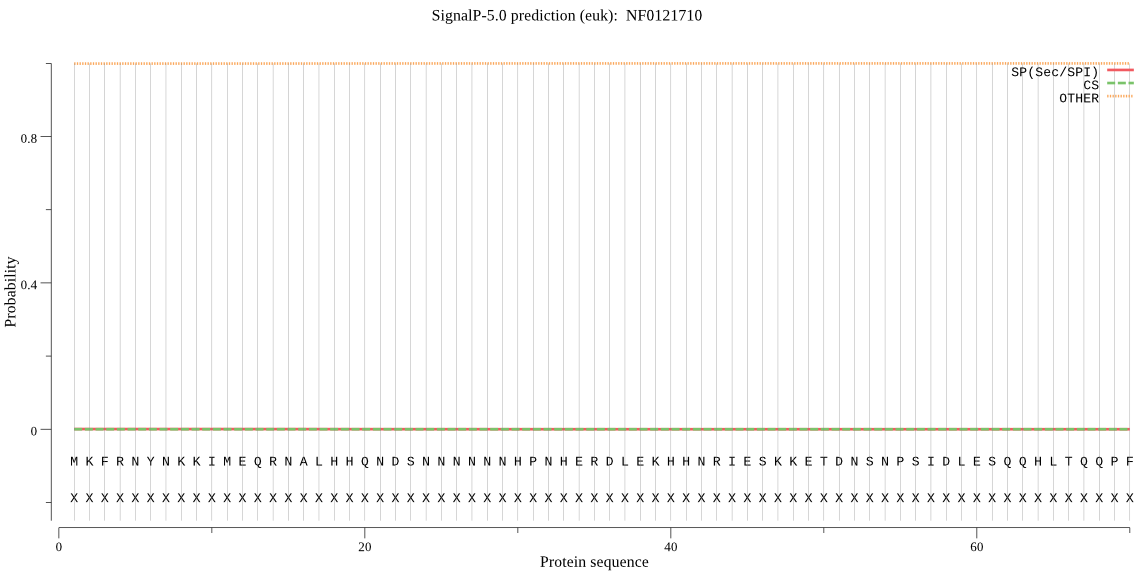
MKFRNYNKKIMEQRNALHHQNDSNNNNNNHPNHERDLEKHHNRIESKKETDNSNPSIDLE SQQHLTQQPFITNDDEQSNLDFYESSFSIFTPLKEEEAPNSHSNIHSSTNTYSSKRTIQS FISSLFSHSITWLIIVSILTVLLWQLPSDVGNYIIYPFTIFGTFWHELGHATTALLCGNT LEFIKIESNGSGLTVYQDVTQNTFCNALISVNGPLGPTFFGCLIICLSVAFVKHELFSRV LLSTIATIVLIVTALFIRKSIFGMVFLPIVSLVLYGIALKAPKVLVTFSLQFIGVQMAIS MYENFMYLFSKMNGKSDTGSVEVLFRGVIPYWLVAILIIVINVTCMTLSLWFVIRHSLRV RQSSSSDEATVVVNQQR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0121710 | 366 S | QSSSSDEAT | 0.997 | unsp | NF0121710 | 366 S | QSSSSDEAT | 0.997 | unsp | NF0121710 | 366 S | QSSSSDEAT | 0.997 | unsp | NF0121710 | 46 S | NRIESKKET | 0.998 | unsp | NF0121710 | 364 S | VRQSSSSDE | 0.998 | unsp |