

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0122010 | OTHER | 0.703488 | 0.140150 | 0.156362 |
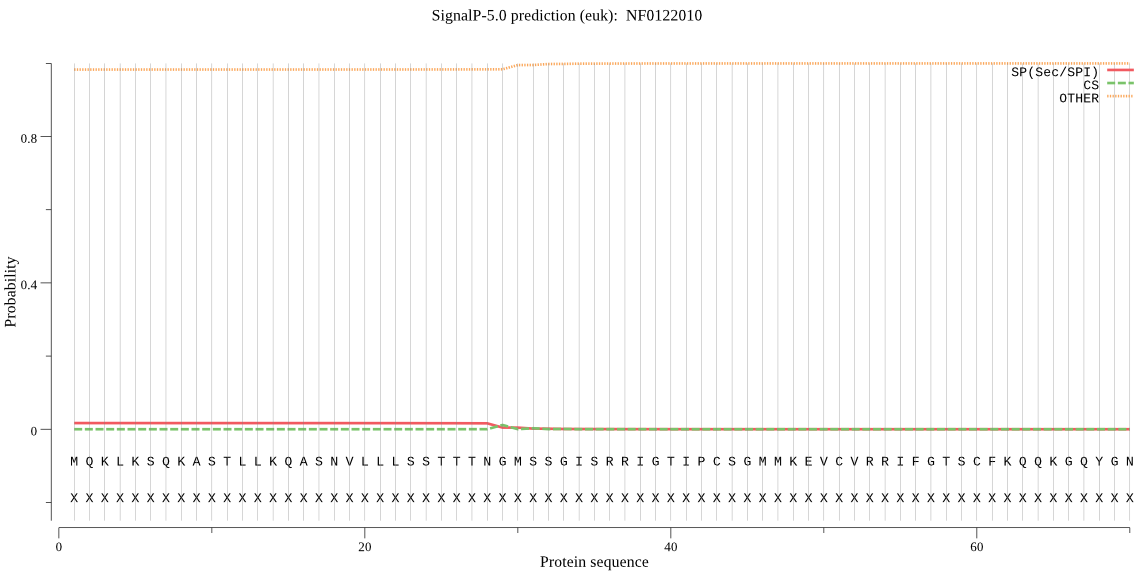
MQKLKSQKASTLLKQASNVLLLSSTTTNGMSSGISRRIGTIPCSGMMKEVCVRRIFGTSC FKQQKGQYGNKTSLMDPNSEIYNLSSSLSASIRLLNQLATQQRAQYHTTNGVFKKKVSVI PSEILNNNILNVVMIVIILIFFNDKMTFRVGY
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0122010 | 6 S | QKLKSQKAS | 0.996 | unsp |