

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0127480 | OTHER | 0.645841 | 0.353918 | 0.000242 |
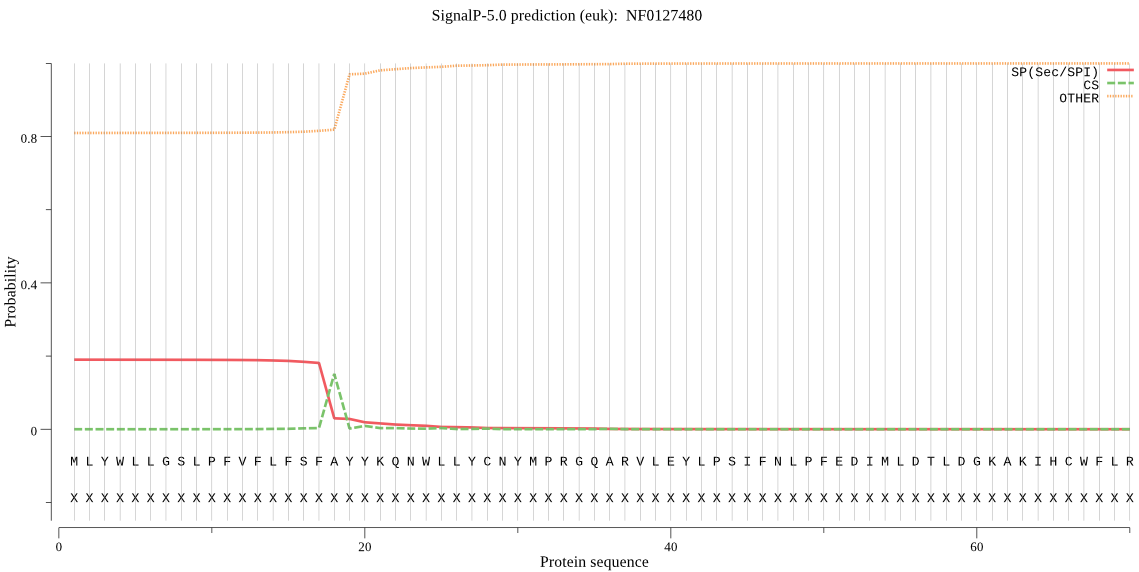
MLYWLLGSLPFVFLFSFAYYKQNWLLYCNYMPRGQARVLEYLPSIFNLPFEDIMLDTLDG KAKIHCWFLRYMPSGGSLDSSFLQHKKSTDKANNDEDAIFSFHHSSSSNATTSSPSSSAS QGNNNSSSGDNLSQQDTKKKIKKGTSSMMLQYKKQLQYTPTIIMFHGNAGNISHRLTNAR DMYRSCKCNILMVEYRGYGRSTGEPSEEGLKNDAETAIRYLLEKRSDINPNNIFIFGRSL GGAVAIFLAAKYANVIRGVIVENTFISVPKLIPSLFPYRFAVPVLQYLCINKWDNETILR SATFRNDFHRKRLNGDLPFLFISGRKDELIPPEHMDTLIEIYCERYGDYCYVKRFEEGTH YGTWLCPGYYLNLYRFIKKFRVNPPSVSETRSRKASSSTTSVKPHIPTSSTDQIHTSGDD IVIHENITLTYADSLTIPPENHHGSGHNSGTSSGANSGDNTPRKKGTYMFR
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0127480 | T | 399 | 0.687 | 0.059 | NF0127480 | T | 400 | 0.667 | 0.027 | NF0127480 | S | 396 | 0.593 | 0.037 | NF0127480 | S | 397 | 0.593 | 0.046 | NF0127480 | S | 398 | 0.573 | 0.049 | NF0127480 | T | 408 | 0.562 | 0.320 | NF0127480 | T | 112 | 0.558 | 0.145 | NF0127480 | S | 401 | 0.557 | 0.081 | NF0127480 | T | 411 | 0.527 | 0.174 | NF0127480 | T | 111 | 0.523 | 0.165 | NF0127480 | T | 390 | 0.523 | 0.032 | NF0127480 | S | 118 | 0.500 | 0.052 |
| ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | ID | Site | Position | Gscore | Iscore | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0127480 | T | 399 | 0.687 | 0.059 | NF0127480 | T | 400 | 0.667 | 0.027 | NF0127480 | S | 396 | 0.593 | 0.037 | NF0127480 | S | 397 | 0.593 | 0.046 | NF0127480 | S | 398 | 0.573 | 0.049 | NF0127480 | T | 408 | 0.562 | 0.320 | NF0127480 | T | 112 | 0.558 | 0.145 | NF0127480 | S | 401 | 0.557 | 0.081 | NF0127480 | T | 411 | 0.527 | 0.174 | NF0127480 | T | 111 | 0.523 | 0.165 | NF0127480 | T | 390 | 0.523 | 0.032 | NF0127480 | S | 118 | 0.500 | 0.052 |
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NF0127480 | 386 S | VNPPSVSET | 0.997 | unsp | NF0127480 | 386 S | VNPPSVSET | 0.997 | unsp | NF0127480 | 386 S | VNPPSVSET | 0.997 | unsp | NF0127480 | 396 S | SRKASSSTT | 0.995 | unsp | NF0127480 | 401 S | SSTTSVKPH | 0.995 | unsp | NF0127480 | 409 S | HIPTSSTDQ | 0.992 | unsp | NF0127480 | 461 T | SGDNTPRKK | 0.99 | unsp | NF0127480 | 133 S | GDNLSQQDT | 0.992 | unsp | NF0127480 | 206 S | TGEPSEEGL | 0.99 | unsp |