

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
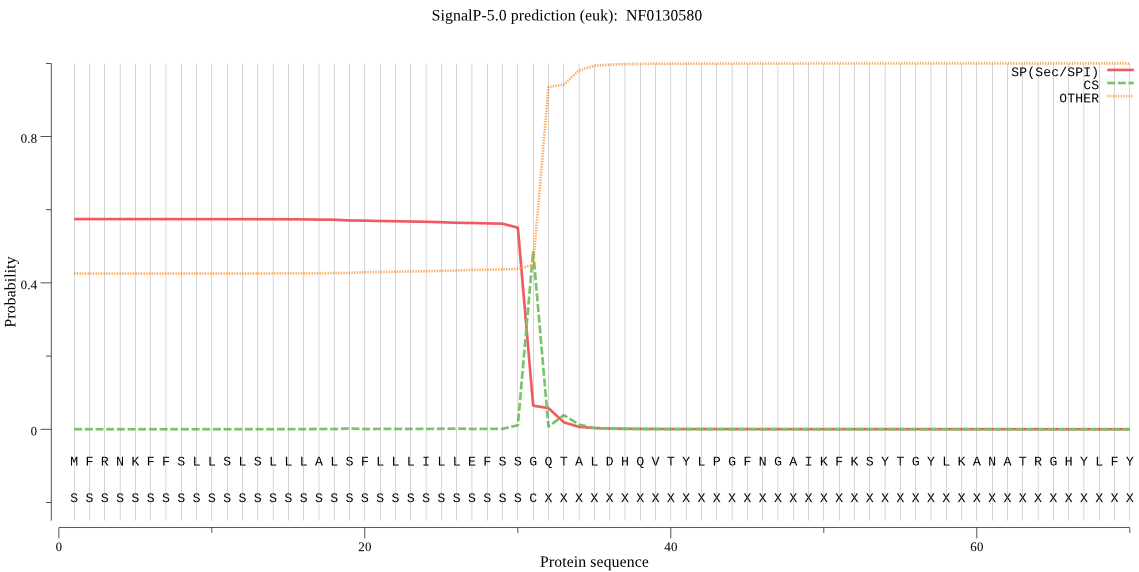
| NF0130580 | SP | 0.040057 | 0.959813 | 0.000130 | CS pos: 31-32. SSG-QT. Pr: 0.9247 |
MFRNKFFSLLSLSLLLALSFLLLILLEFSSGQTALDHQVTYLPGFNGAIKFKSYTGYLKA NATRGHYLFYWFMESQNNPATDPVVWWTNGGPGCSSIDGMVSEHGPFVVLQDGKTVVDNP YAWNKRVNMVFLEQPIGVGYSYSDTPADYQTINDDVAAADMNGAIRDFFTRFPQYLKNDF YIAGESYGGVYVPTAAFRVIQGNQQGEVPKINLKGIMVGNGVTDAESDANSVPLFYKEHS LITAEDYNKGFVACKRNFYANQNVPDCSNFLQLVSSSLNHLNPYYIYDSCTWLGDNGLTK KNSKNHPLFELYKQRPKTKKPFIVGAESDSPCVPDHSIISYFNNPAVRAAIGATHPIASP SGWQVCSTFINYTQIYTTLLPFYSKLLPEIRILAYSGDVDCVLNTSGTKAGLDKLGLNVS LPYTKWIHYDENNDIVVDGFIRKYSNGKGLTFVTVRGAGHMVPQYRPQQALRMLDNFLND KWITKN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| NF0130580 | 141 S | GVGYSYSDT | 0.995 | unsp |