

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| NF0130830 | OTHER | 0.994517 | 0.002790 | 0.002693 |
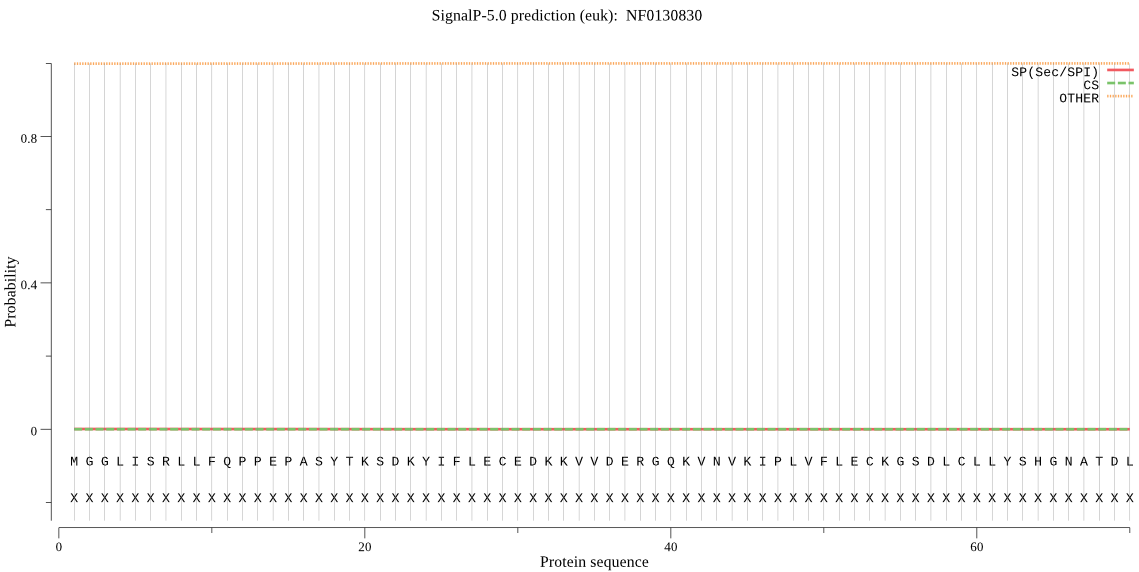
MGGLISRLLFQPPEPASYTKSDKYIFLECEDKKVVDERGQKVNVKIPLVFLECKGSDLCL LYSHGNATDLGQTMPYLELLRSSLKINVCGYEYQGYGISEPKVSCSEPRVYASIEAAVKY LKTERGFSEDRIIVYVLVIRFMIITRDFKYLCDNTNS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| NF0130830 | 104 S | EPKVSCSEP | 0.993 | unsp | NF0130830 | 128 S | ERGFSEDRI | 0.994 | unsp |