

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_0216741 | OTHER | 0.999987 | 0.000009 | 0.000004 |
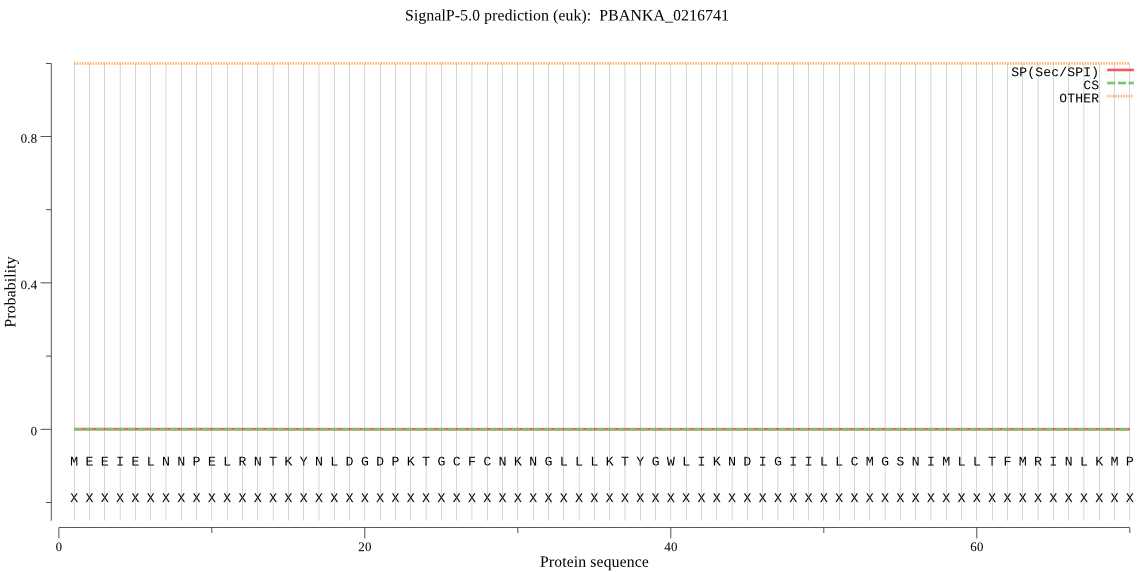
MEEIELNNPELRNTKYNLDGDPKTGCFCNKNGLLLKTYGWLIKNDIGIILLCMGSNIMLL TFMRINLKMPNNNEDLVVGNNNYYIYKDSWIEKFNKMVIQYMHYICKDIGNHKHGKFKRK FNSFDDLVDDVIQYMNQIQDEISNDNQTDDESHNIVTTKKKLPMYIIGHSMGGNIALRLL QLLRKAKEDIIKE*NSNNYKNCNTMLENFTNVNEVAMMNNSNDYNSDNSCASTSATTNTI ASDKDEGFYNYLYKLNIKDCATLFGMMRIKTILNDGKNSFKHFYLPIVNFMSRIATHALI PSKFRYNISKYVASIYKHDNFRNNNGIKFKCMAELIKATIQLECNINYIPNDIPLLFVHS TDDSVCSYKGAV*FYNKANVNEKKLHLVDDMNHDITMGPGNENILKKIIE*ISNLRRDDE DEINGEIEDEKENET
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PBANKA_0216741 | 242 S | NTIASDKDE | 0.997 | unsp |