

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
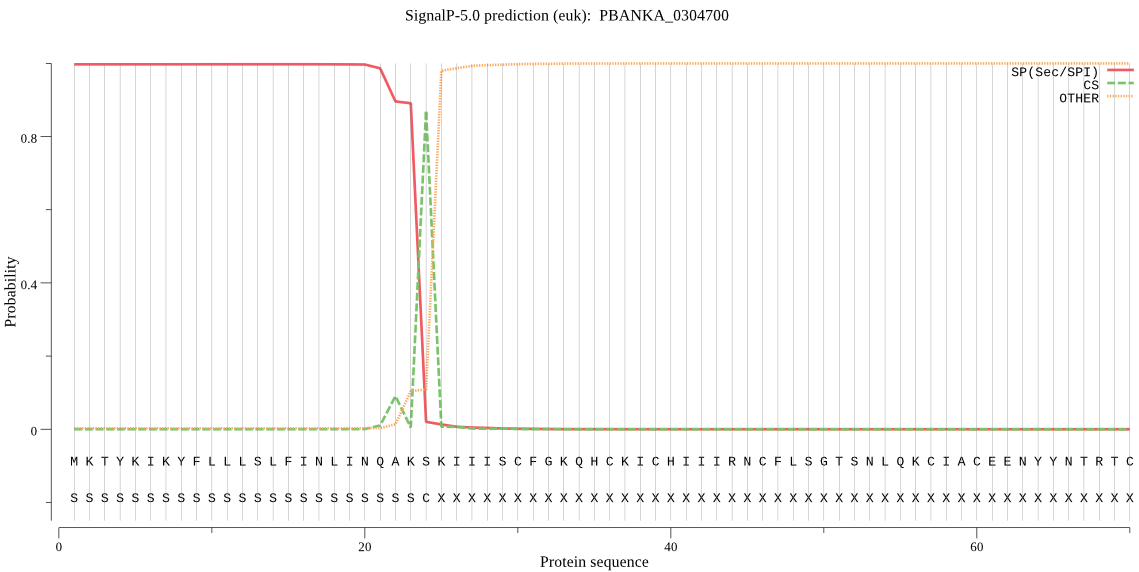
| PBANKA_0304700 | SP | 0.023800 | 0.976166 | 0.000034 | CS pos: 24-25. AKS-KI. Pr: 0.9387 |
MKTYKIKYFLLLSLFINLINQAKSKIIISCFGKQHCKICHIIIRNCFLSGTSNLQKCIAC EENYYNTRTCIKHEESFFNYKGTNALLELRDQSLQDNVSEESLSELISKILDVAIFRYEN KKTDANSMDDDYKSKISNLCLYLNFKDNYESAKNHKQTDVEHIESHIQHIVKTFIHANDN IEHMKNALRNPALCFQNPLEWVQDRLGYKENDEIPSVGIIPEKKLFKPYTNKSLMSSLYN ANSKCNRTYCNRFSDPNECEYNIRPLNQGTCGNCWAFASSTTISAYRCRKGLGFAEPSIK YVTLCKNKYLDDEDSQTFGHYNDNICHEGGHISSYIEILDASKMLPTSFDVPYNEPIKGG ECPTEVSTWGNIWNGVSSLSKILNGYIYKGYFKISFLDYVQAGKTNELINILKDYIIEQG AIFVSMEISQKLNFDHDGEQVMLNCEYGEVPDHALVLIGFGDYIKSSGEKSSYWLIRNSW GSRWGSNGNFKMDMYGPRNCNGRVLFNAFPLLLKMKENNISKPLPNDISSTDVKIRYKHS DFTKNKKNQRDQNRKITPNNYDDPYNFKPKTVDPPNNNNNDYVNPDYNSIDANDRQNNER FYPPSRDPEYDPSYPSMNNNRRFDPSDIINYRNKLRKIFQTDLTVTIGNFVYKRNIYSKR KNEYMEQFSCLRTYSMDSNLDNICRENCEKHIDTCMHSEVIGDCLDRNAPNYKCVYCGM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0304700 | 151 S | DNYESAKNH | 0.993 | unsp | PBANKA_0304700 | 151 S | DNYESAKNH | 0.993 | unsp | PBANKA_0304700 | 151 S | DNYESAKNH | 0.993 | unsp | PBANKA_0304700 | 582 Y | NNNDYVNPD | 0.991 | unsp | PBANKA_0304700 | 93 S | LRDQSLQDN | 0.992 | unsp | PBANKA_0304700 | 127 S | TDANSMDDD | 0.996 | unsp |