

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
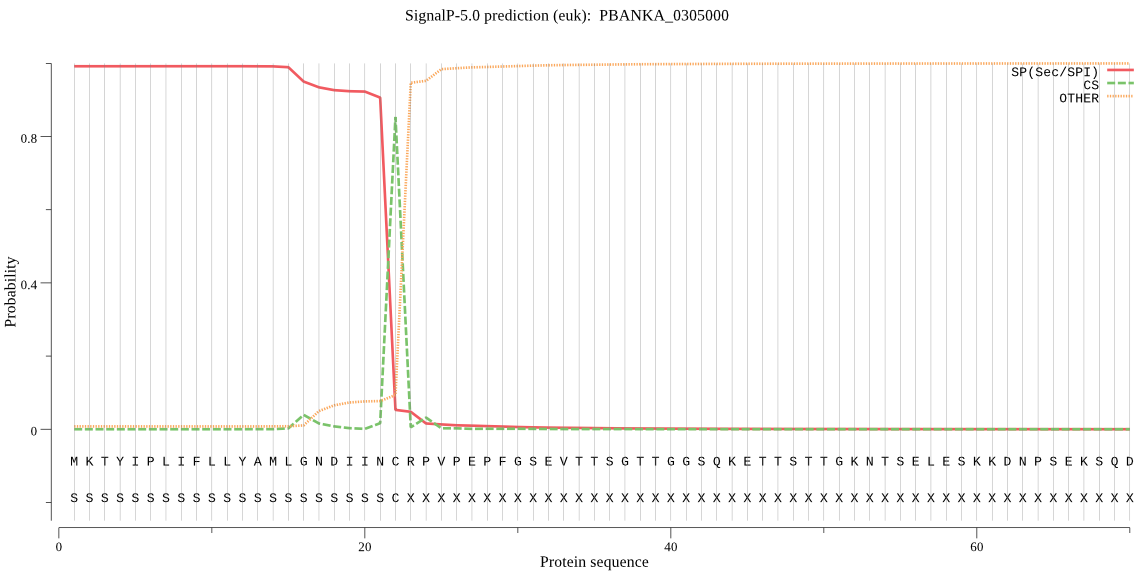
| PBANKA_0305000 | SP | 0.030290 | 0.969710 | 0.000000 | CS pos: 22-23. INC-RP. Pr: 0.8912 |
MKTYIPLIFLLYAMLGNDIINCRPVPEPFGSEVTTSGTTGGSQKETTSTTGKNTSELESK KDNPSEKSQDPSTPSSNTATLPGSPGTSGTPNPSDSSDSSDSSDSSDSSDSSDSSDSSDS SDSSDSSDSSGTTSETPGTPTGTERSLSPQPQNSQPQNSQTQDSQTQDSQTQQQSQRSEA TTGSDSSKNATEEPQSDDASISSNETNVVNNSVIKPRIVHKGTVESALLKNYKGVKVTGL CGDEVGIFLEPHIYINVESKINNIELSAKFPSIHGQYITIFSEESPLKNKCHENDDHLYY KILVYLEGDILTIKWTTFSINPETNKCELCTNRNNENTNVDVKKFRIPKLETPFTTIQIH SLLTKENKIVVKTKDYSLIDDIPKKCDIIATKCFLSGKTDIEPCYTCNLLTENIPRNDEC YNYSSPLVKEYNQILTIGQSDEDNIENQNLSLVDSINNILNDIYIIDEQNNKILIDVEDL NTNLKKELTYYCQILSEVDTSGTLDIYKIGNSVEIFNNLIRLLKNHQNENRLYIINKLKN PAICMKHVEQWVANRKGLKLPIILDDAETENKNLEELNNTNSTINYEEGLDGIIDLTKTI DDEPVSPLGLLANKLDAFCNNEFCDRLKDKTSCVSKIDVEEQGNCATSWIFASKYHLETA ICMKGHDNFNTSALYVANCSKKDPKDKCLSGSNPLEFLDILQANNYLPTESNYPYNYKHV GDSCPETNQTWVNLFDKLKLENSNDGNNSIIKGYTSYESKDYKSNMNEFVGIIKNKIKKL GSVIAYAKFNDFMGYDFNGNKVHKLCGSGTPDMIVNIIGYGKYINENNEVKSYWIVRNSW GKHWADEGNFKVDIETPENCEHNFIHTALVFDFDLPPAQKNNNDASIYNYYLKNSPNFYK NLYYNHVSKTNAETAIFGQDETVQLSNAAGGGSNQVATTINTVTSQLASGATTQINQPST TTQGQAPQSQTCPSSAVTPSQTPSGSSSGSGPTPSGGNNTSHQNNPGQSGASVQPNPSTT VETPENGGGSTERQTNSTQNTEIFHVLKVIRKKKVQTSLIKYNTYDEVEEHSCSRAMSTD SSKKEKCVTFCSNNWDSCSFNLYPGYCLNKLKGDNECNFCAV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0305000 | 59 S | SELESKKDN | 0.998 | unsp | PBANKA_0305000 | 59 S | SELESKKDN | 0.998 | unsp | PBANKA_0305000 | 59 S | SELESKKDN | 0.998 | unsp | PBANKA_0305000 | 65 S | KDNPSEKSQ | 0.997 | unsp | PBANKA_0305000 | 99 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 102 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 105 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 108 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 111 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 114 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 117 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 120 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 123 S | DSSDSSDSS | 0.993 | unsp | PBANKA_0305000 | 148 S | ERSLSPQPQ | 0.993 | unsp | PBANKA_0305000 | 186 S | TGSDSSKNA | 0.997 | unsp | PBANKA_0305000 | 377 S | TKDYSLIDD | 0.994 | unsp | PBANKA_0305000 | 635 S | TSCVSKIDV | 0.99 | unsp | PBANKA_0305000 | 680 S | VANCSKKDP | 0.993 | unsp | PBANKA_0305000 | 756 S | KGYTSYESK | 0.996 | unsp | PBANKA_0305000 | 1030 S | NGGGSTERQ | 0.993 | unsp | PBANKA_0305000 | 1078 S | SRAMSTDSS | 0.992 | unsp | PBANKA_0305000 | 1081 S | MSTDSSKKE | 0.99 | unsp | PBANKA_0305000 | 1082 S | STDSSKKEK | 0.997 | unsp | PBANKA_0305000 | 42 S | TTGGSQKET | 0.997 | unsp | PBANKA_0305000 | 55 S | GKNTSELES | 0.993 | unsp |