

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
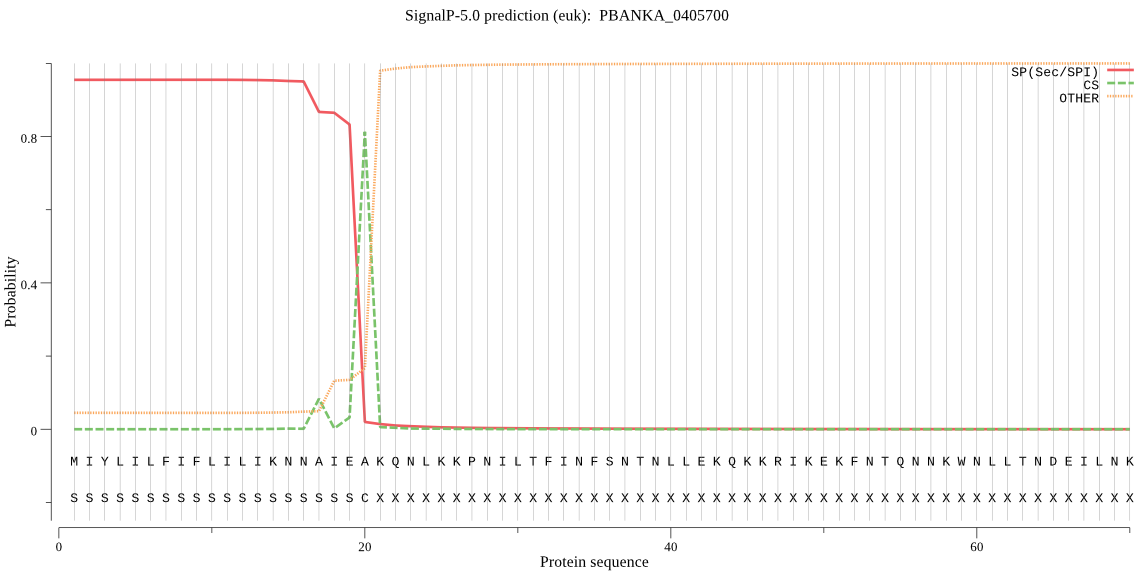
| PBANKA_0405700 | SP | 0.039461 | 0.960216 | 0.000323 | CS pos: 20-21. IEA-KQ. Pr: 0.8470 |
MIYLILFIFLILIKNNAIEAKQNLKKPNILTFINFSNTNLLEKQKKRIKEKFNTQNNKWN LLTNDEILNKNIGVNEINDVKNFSGNLDKTKVNINNVTKIYQEEKSKVRQNVMEKMVEKI LKKKKYKKLEKLLCCNNSIKDMKKNIKLYFFKKRIIYLTDEINKKTSDELIKQLLYLDNI NHNDIKIYINSPGGSINEGLAILDIFNYIKSDIQTISFGLVASMASVILASGKKGKRKSF PNCRIMIHQPLGNAYGHPHDIEIQTKEILYLKKLLYYYLSKFTNKNEETIEKHSDRDYYM NAYEAKEYGIVDEVVETKLTHPYFIGLIKN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0405700 | 167 S | NKKTSDELI | 0.994 | unsp | PBANKA_0405700 | 294 S | IEKHSDRDY | 0.998 | unsp |