

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
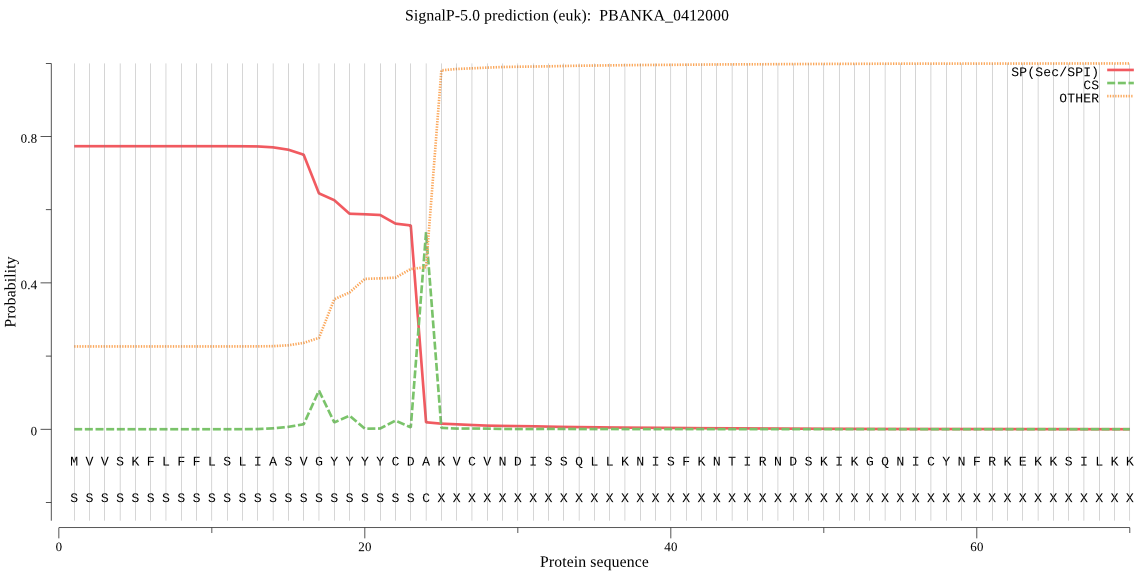
| PBANKA_0412000 | SP | 0.010390 | 0.989580 | 0.000029 | CS pos: 24-25. CDA-KV. Pr: 0.9191 |
MVVSKFLFFLSLIASVGYYYYCDAKVCVNDISSQLLKNISFKNTIRNDSKIKGQNICYNF RKEKKSILKKKIKKKYSLNYSKGCIQKKKKYNLYFLNSSKFTDIKENVKAWFNLDYIENH YAYSYIKSFARIPPITKIYLINTFILSVLIHLNKNVYKYILYDFDKIFKNGEIWRLVTPY FYIGNLYLQYILMFNYLNIYMSAVEIAHYKNPEDFLIFLTYGYISNILFTIIASMYNENV MDLKKYIHKIINLIITIPSNNNKDDKKLVQKETYNHLGYVFSTYILYYWSRINEGSLINC FDLFFIKAEYIPFFFVIQNILLYNEFSFFEVASILSSYTFFTYEKYLKSNFLRDLFRGFL KSIRVYPMYNMYKEEYE
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PBANKA_0412000 | 49 S | IRNDSKIKG | 0.99 | unsp |