

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PBANKA_0517600 | SP | 0.040924 | 0.957882 | 0.001195 | CS pos: 24-25. ILS-IP. Pr: 0.5281 |
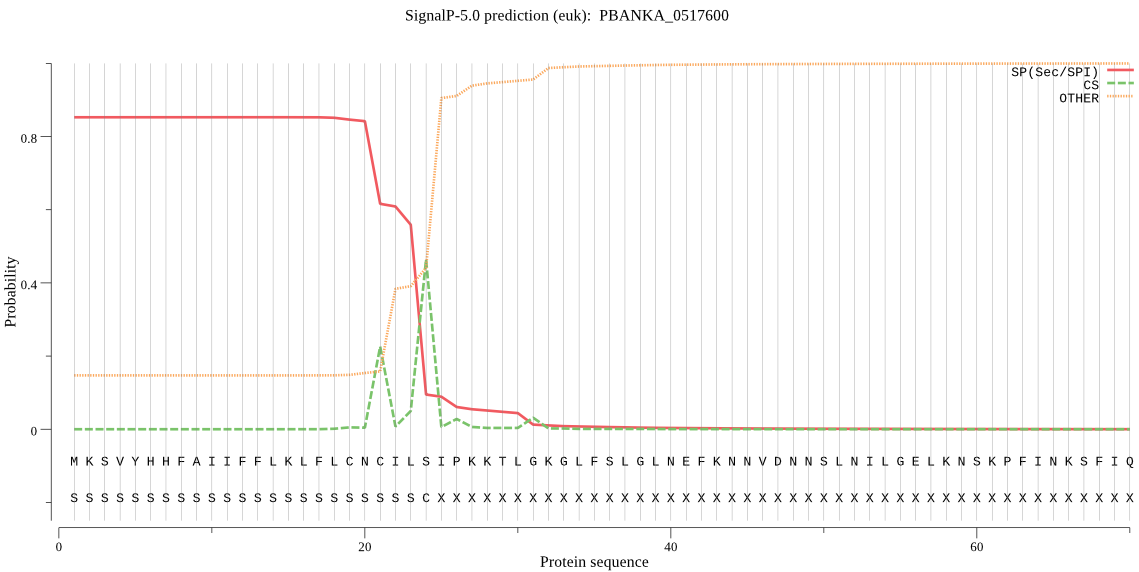
MKSVYHHFAIIFFLKLFLCNCILSIPKKTLGKGLFSLGLNEFKNNVDNNSLNILGELKNS KPFINKSFIQINEKKDNVLLLKLYKQNIASDKLSTYYGKIAIGENSENIFNVLFDTGSTE FWVPFKTCKFTKNNIHNKYERTQSFKYKYDDKGLPSVLEINYLSGKLVGFDGYDTVYLGP GFAIPHTNIAFATSIDIPVLEKFKWDGIIGLGFENEDSQKRGIKPFLDHLKDEKILTEKN YKNIFGYYITNTGGYITLGGIDNRFKRSPDEKIIWSPVSTEMGFWTIDILGIRKEKQPYM NERRDDEVIVKYEGFHDGSNKSIVDTGTFLIYAPKKTIENYLNDLTINSCEDKQKLPYII FQIKSKEIESIKGLSVIELVLSPNDYVIEYIDEVNSTKECIIGIQSDEDNINGWTLGQVF LKSYYTIFDKDNLQIGFVRNKQTINDETYLNESFLRVSKKRNKKKSYNGPL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBANKA_0517600 | 322 S | GSNKSIVDT | 0.993 | unsp | PBANKA_0517600 | 322 S | GSNKSIVDT | 0.993 | unsp | PBANKA_0517600 | 322 S | GSNKSIVDT | 0.993 | unsp | PBANKA_0517600 | 375 S | IKGLSVIEL | 0.991 | unsp | PBANKA_0517600 | 382 S | ELVLSPNDY | 0.994 | unsp | PBANKA_0517600 | 396 S | DEVNSTKEC | 0.994 | unsp | PBANKA_0517600 | 406 S | IGIQSDEDN | 0.993 | unsp | PBANKA_0517600 | 458 S | FLRVSKKRN | 0.997 | unsp | PBANKA_0517600 | 218 S | ENEDSQKRG | 0.992 | unsp | PBANKA_0517600 | 279 S | WSPVSTEMG | 0.993 | unsp |